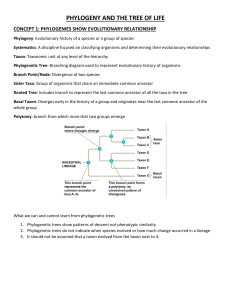
Phylogeny AP Biology Topic 7.9 Phylogenetic Trees and cladograms show the evolutionary relationship of species over lineages. Cladogram These diagrams are hypotheses of evolutionary relationships based on molecular and morphological data. Phylogenetic Tree However, Phylogenetic Trees are calibrated using the fossil record and molecular clocks to show the amount of change. Let’s look at how to read and understand them… Present Time (or more recent) Past Let’s look at how to read and understand them… These represent the different species being studied. Common Ancestor of A, B, and C Point of divergence/ speciation Common Ancestor of A, B, and C The lines represent the series of ancestors over time, leading to the present day species. Unique evolutionary history for species D Shared evolutionary history for species B,C, and D, but not A Shared evolutionary history for all four species How to find Common Ancestors and Relatedness. B and C are the most closely related because they share the most recent common ancestor. Arrow 1 represents the most recent common ancestor of all four species, A-D. Arrow 2 represents the common ancestor of B, C, and D Arrow 3 represents the common ancestor of B and C. B is more closely related to D when compared to A because B and D share a recent common ancestor at #2, which is more recent in terms of time compared to the recent common ancestor at #1, which is further back in time. Rotating the species at the different divergence points, doesn’t change the evolutionary relationships. The distance and relatedness remain the same. Rotating the species at the different divergence points, doesn’t change the evolutionary relationships. The distance and relatedness remain the same. All four of the images below all show the same evolutionary relationships When “reading” a phylogenetic tree or cladogram, the order of the species at the top is not the important part, but rather the locations of the branch points within the tree. The next question we need to ask, is “How do we construct a phylogenetic tree?” Morphological, biochemical, molecular, or behavioral data can be used to construct a tree. DNA or amino acid sequences Most accurate & reliable. By analyzing the data, we can begin to group organisms into different species. Character Table Trait Six legs Blue spots Antenna Wings 1 Species 2 3 4 Outgroup: The lineage that is least closely related to the remainder of the organisms in the phylogenetic tree or cladogram. 1 3 Shared derived traits: Traits that indicate common Wings ancestry 2 4 Antenna Blue dots Six legs 1 3 2 4 Wings Antenna Blue dots Six legs Using Molecular Data to Construct a phylogenetic tree or cladogram Number of Amino Acid Differences in Protein X Species A Species B Species C Species D Species E Species A --- Species B --4 4 --- 2 6 --- 6 1 5 --- 10 11 9 12 --- Species C Species D Species E E Outgroup C A B D E A C B D