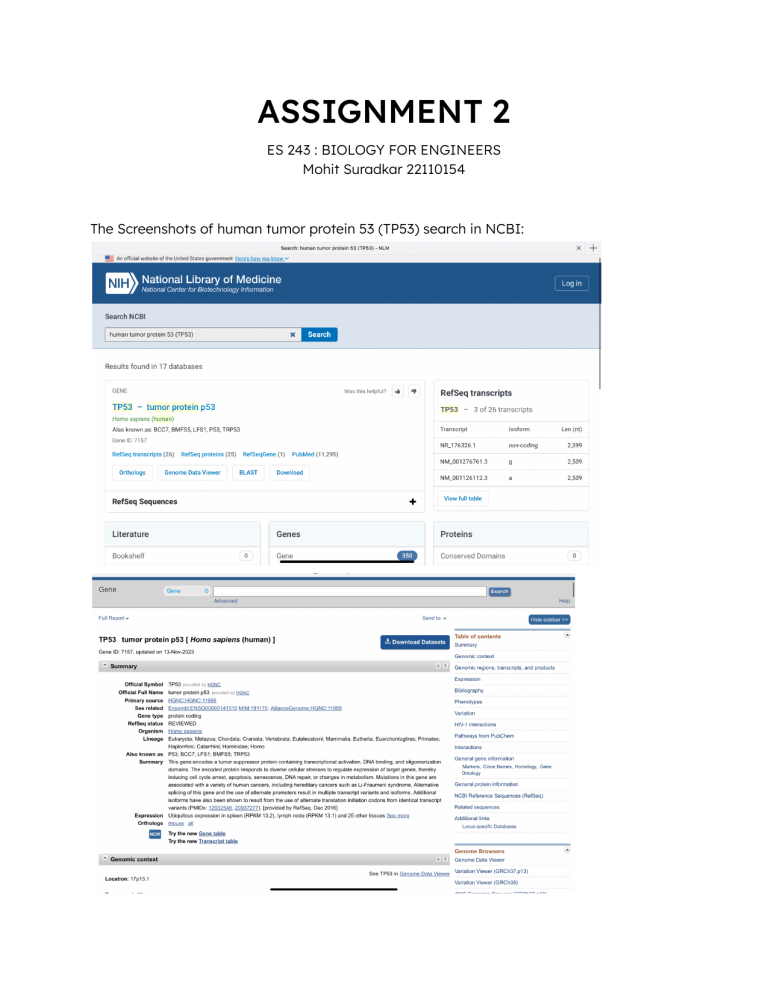
ASSIGNMENT 2 ES 243 : BIOLOGY FOR ENGINEERS Mohit Suradkar 22110154 The Screenshots of human tumor protein 53 (TP53) search in NCBI: Number and Length of Reference Sequences: Reference Gene Sequence (1): NCBI Reference Sequence: NG_017013.2 Length of Sequence : 32772 bp Reference Protein Sequence (25): Some of the Reference sequences and their lengths are listed below: Reference Sequence Length NP_001394197.1 341 aa protein NP_001394198.1 302 aa protein NP_001394200.1 302 aa protein NP_001394196.1 354 aa protein NP_001394194.1 354 aa protein NP_001394195.1 393 aa protein NP_001263628.1 187 aa protein NP_001263627.1 182 aa protein Reference Transcript Sequence (26): Some of the Reference sequences and their lengths are listed below: Reference Sequence Length NM_001276698.3 2,136 bp linear mRNA NM_001276697.3 2,003 bp linear mRNA NM_001276696.3 2,645 bp linear mRNA NM_001276699.3 2,063 bp linear mRNA NM_001276761.3 2,509 bp linear mRNA NM_001276760.3 2,512 bp linear mRNA NM_000546.6 2,512 bp linear mRNA Taking two random Transcript sequences and performing Pairwise Sequence Alignment Using Needleman Wunsch Algorithm Scoring System Used in above : Gap open : 10 Gap extend : 0.5 End open : 10 End extend : 0.5 Gap penalty : 10 Extend penalty : 0.5 End Gap penalty - False Using Smith Waterman Algorithm Scoring System Used in above : Gap open : 10 Gap extend : 0.5 Steps for searching homolog of TP53 protein from House Mouse : Performing pairwise alignment of human and mouse homolog of this protein : Identity : 7775/19516 (39.8%) Similarity : 7775/19516 (39.8%) Gaps : 8467/19516 (43.4%) Score : 33462.5 Structure of TP53 protein : PDB (Protein Data Bank) and FAIR Data Principles: Findable: PDB assigns unique identifiers to protein entries, simplifying structure discovery through advanced search and query options. Accessibility: Unique IDs and rich metadata (experimental method, resolution, authors, etc.) enhance accessibility to PDB data. Interoperability: PDB collaborates with UniProt, Ensembl, ChEMBL, and PDBe-KB, using common formats for seamless data sharing. Reusability: PDB data finds applications in education, drug creation, structural biology, and bioinformatics. NCBI (National Center for Biotechnology Information) and FAIR Data Principles: Findable: Persistent identifiers (Accession numbers, DOIs, PMIDs) and rich metadata aid in efficient data discovery on NCBI. Accessibility: Standard protocols (HTTP, FTP, API) and authentication methods (NCBI account, API key, My NCBI preferences) support easy access to NCBI data. Interoperability: NCBI collaborates with PubMed Central, EMBL-EBI, DDBJ, and UniProt, ensuring interoperability with widely accepted formats. Reusability: NCBI data is reusable for sequence alignment, phylogenetic inference, literature mining, and genomic analysis, with clear data usage licenses (CC0, CC-BY, public domain).