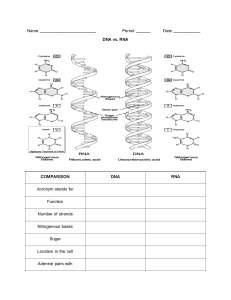
Lecture 5. Genetic Flow of Information: DNA and RNA processing Assignment: Chapter 4 “How Cells Work” - Shuler and Kargi; and for refresher: DNA, RNA and Protein Synthesis reading (blackboard) Definitions: Replication = DNA à DNA Transcription = DNA à RNA Translation = RNA à Protein All DNA in a cell can be replicated (copied) or transcribed (converted to message) I. DNA replication – On your own II. DNA transcription – List steps for DNA transcription A. The central dogma: DNA à mRNA à Protein B. In prokaryotes (single-celled organisms) chromosomal DNA is circular. In eukaryotic cells, DNA is linear, each forming a complex with associating proteins. C. Within this DNA are found individual units of genetic information known as genes. D. Genes can be positioned singly OR can be arranged in clusters known as operons. E. Each gene is transcribed to an RNA molecule, forming either an mRNA (leading to a single protein), a tRNA or an rRNA. tRNA and rRNA are extremely stable species, whereas mRNA is extremely unstable in the cell with a t1/2 = 1 min. F. RNA synthesis is mediated by an enzyme known as RNA polymerase. Again, an enzyme catalyzed process. G. RNA polymerase always reads ANTISENSE strand from 3’ à 5’. Example: 5’ ATGAACGTTTAC 3’ DNA SENSE strand 3’ TACTTGCAAATG 5’ DNA ANTISENSE strand Transcription 5’ AUGAACGUUUAC 3’ mRNA transcript Translation N – Met - Asn - Val - Tyr – C III. DNA information content A DNA sequence encoding a gene(s) has specific pieces of information that govern transcription: A. Promoter = a specific sequence of DNA located near the beginning of the gene (or operon). The promoter is specifically recognized by RNA polymerase. RNA polymerase has different affinities for different promoters, classified as weak or strong promoters. B. Transcription start site = DNA position near the promoter where the mRNA transcript will begin C. Operator = site for additional transcriptional control (discussed later on) D. Transcription terminator = stop signal telling RNA polymerase to terminate. RNA polymerase disassociates from the DNA template and the RNA transcript is released. These features of transcription are universal. However, there are some significant differences in transcription between prokaryotes and eukaryotes that you should be aware of: A. Prokaryotic genes are often arranged in operons without interspacing terminators. Thus, when the RNA polymerase binds, a single polygenic or polycistronic mRNA strand is transcribed. This one long message may result in the subsequent formation of several proteins. Eukaryotic cells do not produce polycistronic messages. B. In prokaryotes, chromosomal DNA is not segregated from the ribosomes so often times the ribosome and protein synthesis will begin even before the entire mRNA has been completely transcribed. In eukaryotes, mRNA transcription is segregated from translation by the nuclear membrane. C. In eukaryotes, mRNA can contain an intervening sequence known as an intron in the middle of the sequence. This intron is cut out of the transcript at two specific sites and the ends of the remaining fragments are joined by a process known as mRNA splicing. IV. Transcriptional control – The lactose operon NOTE: In general, the cell senses when it has too much or too little of a particular protein and responds by increasing or decreasing the rate of transcription of that gene. Background: A. In bacteria, the lactose (or lac) operon controls synthesis of 3 proteins (LacZ, LacY and LacA) needed to utilize the sugar lactose as a carbon and energy source. B. The 3 proteins are encoded by the genes lacZ, lacY and lacA. (SAY SOMETHING ABOUT LOWERCASE ITALICS FOR GENES AND CAPS FOR PROTEINS!) C. lacZ = b-galactosidase which cleaves lactose to glucose and galactose. D. lacY = permease which increases the rate of lactose uptake by pumping lactose into cell. E. lacA = thiogalactosidase transacetylase which degrades small carbon compounds. Organization of genes: lacI (1082) bp—P(84 bp)/O(39 bp)—lacZ (3074 bp)—52bp—lacY(1253bp)—66bp— lacA(611) F. lacZYA is transcribed as a polycistronic message G. RNA polymerase must bind to the promoter/operator region (P/O) in order to initiate transcription. Note that RNA polymerase has low affinity for DNA except in the promoter regions. H. However, if a repressor protein is present in the operator region (O), the polymerase is excluded from binding to the promoter and DNA transcription cannot proceed. LacI is the repressor protein of the lac operon and is called the lac repressor. I. If an activator is present, this can enhance the affinity of RNAP for binding and DNA transcription is massively increased. CAP (catabolite activating protein) is the activator protein. Here’s how it works: Scenario 1: When glucose high, lactose low à LacI represses lac operon, lacZYA OFF A. Bacteria cells normally do not express significant levels of lacZYA because glucose is the preferred food source and LacI is bound to promoter blocking expression. Scenario 2: When glucose low, lactose high à lactose binds to LacI and derepresses lac operon turning on expression of lacZYA genes, lacZYA ON. NOTE: lactose is the signal that induces lacZYA expression. B. When cells encounter a lactose-rich environment, b-galactosidase is needed to convert lactose to metabolizable sugars glucose and galactose. The solution: LacI binds to lactose and changes conformation opening up promoter site for RNAP. LacI is an allosteric enzyme – the binding of lactose to LacI causes a conformational change in LacI that dramatically weakens its affinity for the operator site, allows RNAP to have access. Hemoglobin is another classical allosteric enzyme that changes shape (conformation) when bound to oxygen. C. However, that’s not enough. You also need the CAP activator. CAP is only active when it is complexed to cyclic AMP. cAMP is a hunger signal. When glucose is high, cAMP is low and CAP activator is inactive. When glucose level is low, cAMP level is high. cAMP binds to CAP and the cAMP-CAP complex binds to the operator and causes RNAP to have a 50-fold higher affinity for the lac promoter. Scenario 3: When glucose high, lactose high à Initially, lacZYA is OFF. As glucose decreases, lacZYA turns ON. In summary: A. LacI protein is a REPRESSOR while cAMP-CAP is an ACTIVATOR. Lactose is a primary inducer; cAMP is a secondary inducer. B. This is a classical example of feedback control, used by humans and animals. Talk about throwing a ball at a target with only 3 opportunities – change the later shots based on the first. V. Simple Boolean Logic Many simple genetic networks, including the lac operon, behave as Boolean logic circuits/gates. Refer to handout for details INTRO: A. Boolean networks are logic-based mathematical models commonly used in electrical engineering. B. Gene and protein networks can be described using simple Boolean logic descriptions. C. This is because cellular networks are directed networks – that is they have a well-established sense of direction due to the flow of chemical signals. NODES = Genes or biological stimulus. Note that a biological stimulus can be a chemical signal (inducer), a mechanical force, or a change in temperature or ion concentration. The nodal values represent the level of activity of that node where: high/on = 1 low/off = 0. DISCUSS the NOT, AND, OR, NAND and NOR circuits Note: A NOT gate is also known as an inverter NOT Gate A Q 0 1 1 0 AND Gate A 0 0 1 1 B 0 1 0 1 Q 0 0 0 1 OR Gate A 0 0 1 1 B 0 1 0 1 Q 0 1 1 1 Combination of NOT and AND gates or NOT and OR gates are called NAND and NOR, respectively. These are simple INVERSIONS of AND and OR gates. NAND Gate A 0 0 1 1 B 0 1 0 1 Q 1 1 1 0 NOR Gate A 0 0 1 1 B 0 1 0 1 Q 1 0 0 0 EXAMPLE 1: The Lac Operon – Boolean description A (glucose) 0 0 1 1 glucose A (cAMP) 1 1 0 0 B (lactose/allolactose) 0 1 0 1 Q (lacZYA) 0 1 0 0 cAMP lacZYA lactose/ allolactose