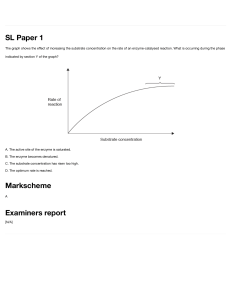
BIOC*2580 Problem Set 5: Answers 1. For 3.3 μmol substrate catalyzed in 5 minutes by 2.0 μg trypsin in a volume of 3.0 mL: Rate of reaction = change in [substrate] or [product] per unit time Change in [substrate] = 𝟑𝟑.𝟑𝟑 𝝁𝝁𝝁𝝁𝝁𝝁𝝁𝝁 𝟑𝟑.𝟎𝟎 𝒙𝒙 𝟏𝟏𝟏𝟏−𝟑𝟑 𝑳𝑳 = 1100 μmol L-1 Rate of reaction = 𝐶𝐶ℎ𝑎𝑎𝑎𝑎𝑎𝑎𝑎𝑎 𝑖𝑖𝑖𝑖 [𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆𝑆] Rate of reaction = 1100 𝜇𝜇𝜇𝜇𝜇𝜇𝜇𝜇/𝐿𝐿 𝑇𝑇𝑇𝑇𝑇𝑇𝑇𝑇 5 𝑚𝑚𝑚𝑚𝑚𝑚 = 220 μmol L-1 min-1 = 2.2 x 10-4 mol L-1 min-1 NOTE: Conversion of mL to L since concertation is expressed as number of moles per litre (3 mL converted to L by multiplying by 10-3) NOTE: μmol is converted to mol by multiplying by 10-6. This is not required unless you are asked to do so (give the answer in molL-1 min-1 instead of μmol L-1min-1) The μ prefix as in μmol indicates micro, 10‐6. Enzyme activity = moles reacted per unit time = 3.3 μmol/5.0 min = 0.66 μmol min-1 (0.66 enzyme units) = 6.6 x 10-7 mol min-1 We have seen the equation: Enzyme Activity = Reaction Rate x reaction volume In this case however, we do not need to use this equation. Since enzyme activity is the number of moles reacted per unit time and the number of moles of substrate that was converted to product is directly given in the question, we can find enzyme activity by: Enzyme Activity (EA) = 𝑁𝑁𝑁𝑁𝑁𝑁𝑁𝑁𝑁𝑁𝑁𝑁 𝑜𝑜𝑜𝑜 𝑚𝑚𝑚𝑚𝑚𝑚𝑚𝑚𝑚𝑚 𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟𝑟 𝑇𝑇𝑇𝑇𝑇𝑇𝑇𝑇 If you use the equation EA = Rate x volume, you will still arrive at the same answer. = 220 μmol L-1 min-1 x (3.0 x 10-3) L NOTE: You MUST convert mL to L = 0.66 μmol min-1 here so that the units cancel out Specific activity = moles reacted per unit time per unit mass of enzyme = Enzyme activity/mass of enzyme present = 0.66 μmol min-1 / 2.0 μg Converting 0.33 μmol min-1μg-1 to mol min-1g-1 = 0.33 μmol min-1 μg-1 = 0.33 mol min-1g-1 0.33 𝜇𝜇𝜇𝜇𝜇𝜇𝜇𝜇 = 0.33 𝑥𝑥 10−6 𝑚𝑚𝑚𝑚𝑚𝑚 -1 -1 min 𝑥𝑥 𝜇𝜇𝜇𝜇 Problem set 5 answers- page 1 1.0 min 𝑥𝑥 1.0 𝑥𝑥 10−6 𝑔𝑔 = 0.33 mol min g Turnover number = activity per mole of enzyme = moles of substrate reacted per unit time per mole of enzyme = molecules of substrate reacted per unit time per molecule of enzyme For trypsin, 1 mol = 25,000 g (Molar mass of trypsin = 25 kDa) Turnover number = specific activity x molar mass of enzyme (with units matched) = 0.33 mol min-1g-1 x 25000 g mol-1 = 8250 min-1 = 138 s-1 NOTE: Converting kDa to gmol-1 25 kDa = 25000 Da 1 Da = 1 gmol-1 25 kDa = 25000 g mol-1 Each trypsin molecule hydrolyses 138 peptide bonds per second. These problems are not mathematically complex except for the need to keep the units straight! 2. From the turnover number and molar mass of catalase, calculate its specific activity. Turnover Number = Specific Activity x molar mass Specific activity = Turnover number/molar mass However, turnover number is usually quoted per second, and enzyme units are usually given per minute. Turnover number of catalase = 5.0 x 105 s-1 Molar mass of catalase = 240,000 g mol-1 Specific activity = 5.0 x 105 x 60 min-1 / (240 x 106 mg mol-1) = 0.125 mol min-1 mg-1 Rate of reaction: Enzyme activity = specific activity x mass of enzyme present Rate of reaction = enzyme activity/reaction volume = 0.125 mol min-1 mg-1 x 1.0 mg of catalase/1.0 L = 0.125 mol L-1 min-1 Problem set 5 answers- page 2 3. a) False: the Michaelis-Menten equation relates variation of the initial rate vo to the value of [S]. However, at very high [S], the dependence approximates to zero order and there is little further increase in vo as its value approaches Vmax. b) False: at saturating levels, i.e. very high [S], vo approaches Vmax and becomes almost constant. c) True d) False: For any given combination of enzyme and substrate, KM is a constant. If an enzyme binds several different substrates, each substrate may have a different KM. e) True for competitive inhibitors, false for non-competitive inhibitors, for which Vmax varies but KM is constant. It's a little doubtful though that any truly non­competitive inhibitors exist - many actual inhibitors have "mixed" inhibition. f) False: This is a common misinterpretation of the statement in part c. KM has units of concentration, Vmax has units of rate, and the two can't be related by a simple numeric ratio. 4. A simple "plug the numbers into the equation" problem: If [S] = 1.0 x 10-4 mol L-1 (= 0.4 KM) then v = 5.8 x 10-7 mol L-1 s-1 If [S] = 2.5 x 10-4 mol L-1 (=KM) then v = 1.0 x 10-6 mol L-1 s-1 (0.5 Vmax) If [S] = 0.5 x 10-4 mol L-1 (= 0.2 KM) then v = 3.3 x 10-7 mol L-1 s-1 If [S] =1.0 x 10-3 mol L-1 (= 4 KM) then v = 1.6 x 10-6 mol L-1 s-1 KM has units of concentration since KM is the substrate concentration that gives rise to 50% occupancy of enzyme, i.e. [ES] = 0.5[E]t, and hence vo = 0.5 Vmax. KM is also by definition (k2 + k-1)/k1; if each rate constant is given its appropriate units, cancellation leaves KM with the units of mol L-1. 5. The two substrate concentrations are "very high", 0.01 mol L-1 = 1000 x KM and 0.001 mol L-1 = 100 x KM respectively. At these concentrations, vo is almost equal to Vmax. "High" and "low" substrate concentration should be judged relative to KM, and KM is very low in this example. A substrate concentration of 0.001 molL-1 would be considered "low" relative to an enzyme with KM = 0.01 mol -1. Problem set 5 answers- page 3 6. Calculation of relative rate vo/Vmax, given [S] relative to KM. Arrange the Michaelis Menten equation in the form, vo/Vmax = [S]/( KM + [S]) Given [S]/ KM = 0.01, then [S] = 0.01 x KM Now write 0.01 x KM in place of [S] in the equation, vo/Vmax = 0.01 x KM / KM (1 + 0.01) cancel the KM from the fraction, vo/Vmax = 0.01/(1 + 0.01) and turn the resulting fraction into a decimal, vo/Vmax = 0.0099 Given [S]/ KM = 0.1, then [S] = 0.1 x KM, vo/Vmax = 0.1/(1 + 0.1) and turn the resulting fraction into a decimal, vo/Vmax = 0.091 Given [S]/ KM = 0.25, vo/Vmax = 0.25/(1 + 0.25) = 0.20 Given [S]/ KM = 0.5, vo/Vmax = 0.5/(1 + 0.5) = 0.33 Given [S]/KM = 1.0, vo/Vmax = 1/(1 + 1) = 0.50 Given [S]/ KM = 2.0, vo/Vmax = 2.0/(1 + 2.0) = 0.67 Given [S]/ KM = 4.0, vo/Vmax = 4.0/(1 + 4.0) = 0.80 Given [S]/ KM = 10, vo/Vmax = 10/(1 + 10) = 0.91 7. To calculate the ratio [S]/KM when vo/Vmax is given, write the Michaelis-Menten equation as: vo/Vmax = [S]/(KM + [S]) put the given value on the left and solve: If vo/Vmax = 0.10, 0.10 = [S]/(KM + [S]) 0.10 x KM + 0.10 x [S] = [S] 0.10 x KM = (1.0 - 0.10) [S] Problem set 5 answers- page 4 divide by (1.0 - 0.10) and divide by KM 0.10/0.90 = [S]/ KM = 0.11 If vo/Vmax = 0.125, 0.125 = [S]/(KM + [S]) 0.125 x KM + 0.125 x [S] = [S] 0.125/0.875 = [S]/ KM = 0.142 If vo/Vmax = 0.333, 0.333 = [S]/(KM + [S]) 0.333 x KM + 0.333 x [S] = [S] 0.333/0.667 = [S]/ KM = 0.50 If vo/Vmax = 0.50, 0.50 = [S]/(KM + [S]) 0.50 x KM + 0.50 x [S] = [S] 0.50/0.50 = [S]/ KM = 1.0 If vo/Vmax = 0.667, 0.667 = [S]/( KM + [S]) 0.667/0.333 = [S]/ KM = 2.0 If vo/Vmax = 0.80, 0.80 = [S]/( KM + [S]) 0.80/0.20 = [S]/ KM = 4.0 If vo/Vmax = 0.90, 0.90 = [S]/(KM + [S]) 0.90/0.10 = [S]/ KM = 9.0 If vo/Vmax = 0.99, 0.99 = [S]/(KM + [S]) 0.99/0.01 = [S]/ KM = 99 (Note that vo/Vmax can never be greater than 1.0) 8. To determine Vmax and KM, you need to do a Michaelis Menten plot. You need to test at least one value of [S] high enough to see the flattening of the Michaelis­Menten curve where [S]>>KM, or you will be unable to estimate Vmax. You need to test at least one value where [S]< KM, or you won't be able to estimate KM. Start by assuming KM is high and try [S] = 10-2 M; with luck this will be at least 10 x KM, and the observed rate will be close to Vmax. Next try a very low [S], e.g. [S] = 10-7 M; with luck this will be lower than the minimum guesstimate for KM. If our luck is good, two very different rates should be observed since the concentrations are far apart, and we can use the remaining four tests to fill in the intervening values between these two extremes. If however two similar rates are observed, we may have an enzyme with an unusually low KM, and even our low substrate concentration sample is giving a rate close to Vmax. Both our measurements are in the flat part of the Michaelis‐Menten plot. In this case, the remaining four tests should be done with even lower [S], until we find a value of [S] that reduces rate to well below half the previously observed Vmax. This will indicate that we have a point on the plot which will allow us to estimate KM. Once we are satisfied we Problem set 5 answers- page 5 have results in the right range, the data should be re- plotted in linear form, e.g. double reciprocal plot, to actually estimate KM and Vmax by graphical means. 9. k2 is the turnover number of the enzyme: Turnover number = molecules of substrate catalyzed per molecule of enzyme per second = moles of substrate catalyzed per mole of enzyme per second Vmax = k2[E]total, i.e. the rate of catalysis when 100% of the available enzyme is occupied. KM is defined as (k-1 + k2)/k1. Many enzymes have a fast substrate binding reaction (the initial reversible step) and a much slower second catalytic step so that k-1 >> k2. KM then approximates to k-1/k1, which is the dissociation equilibrium constant of the ES complex. Thus KM indicates how tightly the substrate is bound (lower KM means tighter binding). Problem set 5 answers- page 6