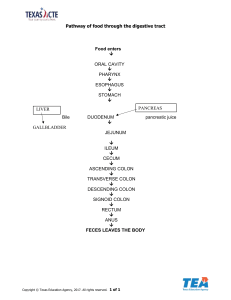
Isolation, identification and characterization of cancer stem cells from different histopathological grades of colorectal carcinoma Aditi Bhattacharya 1 INTRODUCTION • • • According to Globocan reports (2018): – Colorectal cancer (CRC) – fourth most common cancer worldwide. – Second most common cancer in women. – Third most common cancer in men. In India, about 50,982 cases reported in 2015 and the projected numbers for year 2025 is 62,545. (Asia Pacific J of Cancer Prev, 2015) Lifestyle and dietary factors – primary causes for increased incidence of nearly two-thirds of all CRCs. (Gupta S et al. Colorectal Dis., 2010) • Symptoms – fever, abdominal pain, altered bowel habits, fecal blood. • Diagnosis – X-ray, colonoscopy, MRI. • Treatment – Surgery ± adjuvant chemotherapy/radiotherapy. (cancernetwork.com) STAGES & GRADES OF COLORECTAL CANCER TNM Staging Histopathological grading Well differentiated Poorly differentiated Staging : Progression or spread Grading: Cell differentiation Moderately differentiated Normal colon (Kuepper et al., 2016) 3 Tumor Heterogeneity – Origin of Cancer Stem Cell Tumor growth Tumor growth No tumor (www.eurostemcell.org) Cancer stem cell properties Cancer stem cells share a number of characteristics with normal stem cells. NORMAL Stem Cell (kirshner.bio.purdue.edu) Cancer stem cell niche (Anderson E. et al. 2011) 6 Colorectal cancer stem cell markers MARKER FUNCTION ROLE IN TUMORIGENESIS REFERENCES CD 44 (H-CAM) Cell adhesion Hyaluronic acid receptor Colony formation, xenograft growth, Prognostic factor, Correlation with stage Barker et al. 2009, Vaiopoulos et al. 2012 CD 166 (ALCAM) Cell adhesion Prognostic factor Lugli et al.2010 CD 29 (β1- Integrin) Cell adhesion Colony formation Vermeulen et al. 2008 Lgr5 Cell signalling Wnt target gene, crypt base restriction Barker et al. 2007 CD 24 (HSA) Cell adhesion Clonogenic ability, correlation with invasiveness & survival. Vermeulen et al. 2008 Choi et al. 2009 CD 133 (Prominin1) Cell motility Prognostic factor Lugli et al.2010 Elsaba et al. 2010 CD 326 (EpCAM) Cell adhesion Tumor budding & metastasis Gosens et al. 2007 ALDH1 Detoxyfying enzyme Tumor initiation in xenografts, resistance to alkylating agents Vaiopoulos et al. 2012 Targeting of cancer stem cells (Todaro et al., 2010) 8 AIM To isolate, identify, culture and characterise cancer stem cells from different histopathological grades of primary untreated colorectal cancers. 9 OBJECTIVES 1. To isolate, identify and culture CSC from different histopathological grades of primary untreated fresh colorectal cancer tissue. 2. To examine genome wide transcriptomic changes in colorectal cancer stem cells. 3. To validate the putative differential display of genes that help in characterizing different CSC populations. 10 PATIENT SAMPLE CRITERIA • • • • • • • • • • PATIENT INCLUSION CRITERIA: Primary, untreated colorectal cancer cases. Sporadic cases with confirmed histopathological diagnosis. Surgically resectable tumors. No other associated serious medical or surgical conditions. Patients who are willing to participate with informed consent & Institute Ethics Committee approval. PATIENT EXCLUSION CRITERIA: Patients who have received any form of treatment earlier for colorectal cancer e.g. neoadjuvant chemotherapy or radiotherapy. Presence of secondaries in colon or rectum. Patients with family history of colorectal cancers or any hereditary or genetic associations Patients with any other serious medical or surgical condition. Patients who are not willing to give consent for participation in this study. 11 WORK PLAN FRESH NORMAL COLORECTAL TISSUE [from adjoining area] FRESH COLORECTAL CANCER TISSUE [n=6 for each histopathological grade] SINGLE CELL SUSPENSION using Collagenase IV FACS CANCER STEM CELLS TUMOR SPHERE ASSAY BULK TUMOR CELLS NORMAL COLON STEM CELLS NORMAL COLON CELLS RNA ISOLATION MICROARRAY REAL TIME PCR IMMUNOHISTOCHEMISTRY 12 RESULTS Objective 1 To isolate, identify and culture CSC from different histopathological grades of primary untreated fresh colorectal cancer tissue. 13 Clinico-pathological characteristics of patients Age (in years) ≥60 33% Sex ≤30 9% a. Mucinous 6% PD 6% d. Others 12% b. Not known 17% T1 1% M-Stage Not known 17% N2 14% T3 54% Left colon 16% N-Stage T2 18% T4 10% e. Right colon 54% c. T-Stage WD 13% MD 59% Transverse colon 9% Male 61% Tumor Grade SRC 4% Rectum 9% Female 39% 31-59 58% Tumor location Both colon & rectum 12% f. Not known 17% M0 31% N0 51% N1 18% g. Mx 44% M1 8% 1A. SINGLE CELL SUSPENSION Tumor fragment minced into multiple fragments 200 U Collagenase IV per ml of HBSS Incubation for 30-60 min at 37oC Tissue Cell suspension in HBSS Pass through 70 µm filters Pass through 40 µm filters Single cell suspension Cell Counting and staining 15 1B. FLOW CYTOMETRY AND CELL SORTING DATA Colorectal cancer : CD44-PE & CD166-Alexa Fluor647 Normal colon : CD29-PE & Lgr5-Alexa Fluor647 16 Representative flow cytometry analysis of moderately differentiated (MD) CRC and its adjacent normal tissue. 1.3 74.2 21.0 CD44 - PE 3.5 CD166 – AF647 Colon cancer stem cells (DP) – 1.3 %; Bulk tumor cells (DN) – 74.2% 6.6 45.3 0.3 CD29 - PE 47.8 Lgr5 – AF647 Normal colon stem cells (DP) – 6.6%; Bulk normal colon cells (DN) – 45.3 % Representative flow cytometry analysis of well differentiated (WD) CRC and its adjacent normal tissue. 0.7 82.7 4.0 CD44 - PE 12.6 CD166 – AF647 Colon cancer stem cells (DP) – 0.7 %; Bulk tumor cells (DN) – 82.7% 1.3 52.7 0.1 CD29 - PE 46.0 Lgr5 – AF647 Normal colon stem cells (DP) – 1.3 %; Bulk normal colon cells (DN) – 52.7 % Representative flow cytometry analysis of poorly differentiated (PD) CRC and its adjacent normal tissue. 4.9 55.2 1.6 CD44 - PE 38.3 CD166 – AF647 Colon cancer stem cells (DP) – 4.9 %; Bulk Tumor cells (DN) – 55.2 % 1.2 24.2 0.1 CD29 - PE 74.6 Lgr5 – AF647 Normal colon stem cells (DP) – 1.2 %; Bulk normal colon cells (DN) – 24.2 % n=3 *** -- P-value < 0.0001 Dual positive cells isolated from CRC (MD) formed larger and more number of spheres as compared to the dual positive cells isolated from adjacent normal colon tissue. 20 Objective 2 To examine genome wide transcriptomic changes in colorectal cancer stem cells. 21 QC of RNA – Integrity of RNA Samples loaded Gel loaded into chip Ladder Representative Electropherogram obtained by a Bioanalyzer RIN score calculated by Agilent 2100 Expert software 22 2A. RIN SCORE REPORT Well differentiated Tumor P49 – Tumor DP P57 – Tumor DN * RIN score threshold for microarray is ≥ 7.0 P49 – Tumor DN P58 – Tumor DN Moderately differentiated Tumor P45 – Tumor DP P28 – Tumor DP P53 – Tumor DP P45 – Tumor DN P28 – Tumor DN P53 – Tumor DN Adjacent normal tissue P25 – Normal DP P34 – Normal DP P37 – Normal DP P30 – Normal DN P34 – Normal DN P37 – Normal DN 2B. MICROARRAY Isolation of total RNA from tissues/cell samples Preparation of cDNA Transcription, Incorporation of fluorescent label (Cy3) and Amplification of cRNA Fragmentation and charging on the slide (4x44k) Overnight Hybridization Washing of Slide Scanning of slides Feature Extraction Import of data into Analysis Software in txt Format Data Analysis and Derivation of Biological Meaning Principle Component Analysis based on differentially expressed genes between CSCs and NSCs (MD-DP vs N-DP) GeneSpring 14.9.1 *n= 3 NSC CSC Volcano Plot of Differential Expression Analysis Cut off parameters: • Fold change > 2 • p-value < 0.05 Cancer Stem Cells (CSCs) versus Normal Stem Cells (NSCs) Hierarchical Clustering Analysis based on differential expression of genes NSC CSC Differentially regulated signaling pathways S. No. Pathways Targets 1. NFkB pathway 5 2. Chemokine-mediated signaling pathway 7 3. BMP pathway 5 4. Non-canonical Wnt pathway 3 5. Tyrosine phosphorylation 4 6. Positive regulation of PI3K 5 7. Canonical Wnt pathway 8 30 Differential Expression Analysis after Gene Ontology NSC CSC Network Analysis of target genes using GeneGO MetaCore Network analysis uses a complex algorithm which computes the putative interactions between different molecules on the basis of statistical probability. Network Analysis of target genes using GO MetaCore Network analysis confirms that our target genes conform to the signaling pathway. Pathway analysis of differentially regulated genes using GeneGO MetaCore Target genes interact with each other and directly or indirectly contribute towards colorectal cancer pathogenesis. Objective 3 To validate the putative differential display of genes that help in characterizing different CSC populations. 35 Wnt canonical and non-canonical signaling pathway Wnt canonical Wnt non-canonical (Chae et al., 2018) 36 KEGG database Canonical Pathway Planar cell polarity pathway (PCP) Non canonical Pathway 3A. Real-Time PCR Isolation of total RNA from sorted cells from patients Quantify amount of RNA isolated Prepare cDNA using random hexamers Run real time PCR by loading 10 ng cDNA/well Calculate relative gene expression (RGE) by ΔCt method Validation of gene expression by real-time PCR n=15/13 p-value=0.1285 n=15/13 p-value=0.0099 Wnt2 Frizzled10 Β-Catenin LEF1 Wisp1 n=15/10 p-value=0.0073 n=15/9 *Gene expression normalized to 18s rRNA expression *Mann-Whitney U-test p-value=0.0736 (** -- p-value<0.01) 39 Validation of gene expression by real-time PCR p-value=0.1796 n=15/12 n=15/13 p-value=0.3109 Wnt5a Frizzled10 NfatC3 n=15/9 p-value=0.6764 Gene expression of Wnt canonical pathway associated genes (Wnt2, β-catenin, LEF1, Wisp1) and non-canonical pathway associated genes (Wnt5a, NfatC3, Fzd10) were found to be higher in CSC as compared to NSC but only LEF1 and β-catenin showed statistically significant increase. 40 Wnt non-canonical signaling pathway • Wnt5A (Wnt family member 5A) – Regulates osteogenesis in MSCs – Promotes EMT via β-catenin pathway in NSCLC, bladder cancer, breast cancer, prostate cancer, colorectal etc. – Upregulates migration and invasion in osteosarcoma via SRC/ERK/MMP-14 pathway • NfatC3 (Nuclear Factor Of Activated T Cells 3) – Heterodimeric transcription factor – Critical for human postnatal myogenesis, acts via Calcineurin pathway – Upregulated under hypoxic conditions via Orai1Notch pathway in breast cancer. – Involved in SFRP2-induced angiogenesis in endothelial cells. – Associated with high β-catenin expression in human acute infarction tissues, mediates hypertrophy via MAPK pathway (Chae et al., 2018) 41 3B. Immunohistochemistry Deparaffinization of tissue sections Antigen retrieval for 20 mins in citrate buffer Blocking in Peroxide Block Superblock (kit) incubation for 10 mins Primary antibody incubation overnight at 4⁰C Secondary antibody incubation for 1 hour at RT DAB staining and hematoxylin counterstain Coverslip mounting with DPX Analysis: % positivity and H score Wnt5a (1:1000) NfatC3 (1:100) CD44 (1:600) Lgr5 (1:400) Immunohistochemical analysis of Wnt5a expression in CRC (MD) and normal colon tissue sections TUMOR a. b. NORMAL COLON n=31 c. d . Intra-tumoral heterogeneity in Wnt5a expression in CRC (MD) tissue section TUMOR a. b. c. d. TUMOR Immunohistochemical comparison of Wnt5a expression in Epithelium of CRC (MD) and normal colon tissue sections ** WNt5a_Epi_H score WNt5a_Epi_%positivity ** Tumor Normal Tumor Normal n=31 (** -- p-value<0.01) Wnt5a expression was significantly higher in the epithelial region of CRC (MD) tissue as compared to normal colon tissue. Immunohistochemical comparison of Wnt5a expression in intra-tumoral/stromal region of CRC (MD) and normal colon tissue sections ns WNt5a_IT_H score WNt5a_IT_%positivity ns Tumor Normal Tumor Normal n=31 There was no statistically significant difference in Wnt5a expression in the intratumoral region of CRC (MD) tissue as compared to normal colon tissue. ROC curves to understand the sensitivity and specificity of Wnt5a expression in CRC (MD) and normal colon tissue sections •Positivity: AUC 0.613, p-value 0.127, For a positivity of 82.5% or more, sensitivity = 61.3%, specificity = 51.6% •H Score: AUC 0.609, p-value 0.139, For H score of 9.5 or more, sensitivity = 80.6%, specificity = 41.9% •Positivity: AUC 0.631, p-value 0.071, For a positivity of 82.5% or more, sensitivity = 64.5%, specificity = 48.4% •H Score: AUC 0.535, p-value 0.074, For H score of 9.5 or more, sensitivity = 32.3%, specificity = 74.2% 47 Immunohistochemical analysis of NfatC3 (nuclear) expression in CRC (MD) and normal colon tissue sections TUMOR a. b. NORMAL COLON n=31 c. d. Immunohistochemical comparison of NfatC3 (nuclear) expression in epithelium of CRC (MD) and normal colon tissue sections *** a. NfatC3_Epi_H score NfatC3_Epi_%positivity ** Tumor Normal b. Tumor Normal n=31 (** -- p-value<0.01) (*** -- p-value<0.001) NfatC3 expression was significantly higher in the epithelial region of normal colon as compared to CRC (MD). Immunohistochemical comparison of NfatC3 (nuclear) expression in intratumoral/stromal region of CRC (MD) and normal colon tissue sections nsns NfatC3_IT_H score NfatC3_IT_%positivity nsns Tumor Normal Tumor Normal n=31 There was no statistically significant difference in NfatC3 (nuclear) expression in the intra-tumoral region of CRC (MD) tissue as compared to normal colon tissue. ROC curves to understand the sensitivity and specificity of NfatC3 expression in CRC (MD) and normal colon tissue sections •Positivity: AUC 0.728, p-value 0.009, For a positivity of 55% or more, sensitivity = 65.2%, specificity = 77.3% •H Score: AUC 0.769, p-value 0.002, For H score of 7 or more, sensitivity = 65.2%, specificity = 77.3% •Positivity: AUC 0.653, p-value 0.078, For a positivity of 72.5% or more, sensitivity = 65.2%, specificity = 68.2% •H Score: AUC 0.631, p-value 0.131, For H score of 9.5 or more, sensitivity = 65.2%, specificity = 68.2% 51 Immunohistochemical analysis of Lgr5 protein expression in normal colon tissue sections NORMAL COLON a. b. NORMAL COLON n=31 c. d. 52 Immunohistochemical analysis of CD44 protein expression in CRC (MD) tissue sections TUMOR a. b. TUMOR n=31 c. d. Correlation between expression of Wnt5a with NfatC3, CD44 and Lgr5 Correlation coefficient, r= 0.043 p-value=0.37 Correlation coefficient, r= -0.092 p-value=0.311 Correlation coefficient, r= 0.077 p-value=0.341 • There was no correlation between Wnt5a and NfatC3 expression in tumor and normal colon sections. • No correlation was found between Wnt5a and Lgr5 expression in normal colon and Wnt5a and CD44 expression in CRC (MD) sections. 54 Correlation between expression of NfatC3 with Lgr5 and CD44 Correlation coefficient, r= -0.109 p-value=0.28 Correlation coefficient, r= 0.001 p-value=0.497 No correlation was found between NfatC3 and Lgr5 expression in normal colon and NfatC3 and CD44 expression in CRC (MD) section. 55 Summary ➢ The proportion of CSCs present in CRC is variable and differs from patient to patient, even with those having the same histopathological stage and grade. ➢ The percentage of NSCs in normal colon tissue is less variable and varies between 1.0 to 2.5%. ➢ CSCs exhibit a higher proliferation rate as compared to NSCs. ➢ NSCs are potentially quiescent in nature and a much less proportion of cells show sphere-forming ability as compared to CSCs. ➢ Protein localization by IHC of Wnt5a showed positive expression across both epithelium and intra-tumor/mucosal stroma. However, the intensity of Wnt5a expression in tumor was significantly higher than in normal colon sections. ➢ IHC of NfatC3 revealed significant decrease in its nuclear localization and expression, in the epithelium as well as intra-tumor stroma, as compared to normal colon. ➢ Differential gene expression of CSCs derived from moderately differentiated CRCs and their adjacent NSCs reveal regulation of some interesting signaling pathways associated with tumorigenesis like PI3K signaling and BMP signaling pathway. ➢ No correlation could be established between the Wnt5a and NfatC3 expression as well as with established CSC (CD44) and NSC markers (Lgr5). 56 Conclusion NfatC3 acts a potential negative regulator of colorectal cancer. Further study of the gene function will shed light on its role in cancer and cancer stem cells biology. 57