A Brief Overview of Spectral Graph
Convolutional Network
Min Li
Computational Social Sciences and Humanities (CSSH),
Rheinisch-Westfälische Technische Hochschule Aachen,
Templergraben 55, 52062 Aachen, Germany
Abstract. The success of Deep Learning has inspired the development
of learning methods on non-Euclidean domains. Recently, many approaches
have emerged, especially the spectral methods based on spectral graph
theory. In this paper, we provide a brief overview of the spectral methods,
focusing on the fundamental theories and algorithms of three spectral
methods.
Keywords: graph neural network · graph convolutional network · spectral methods.
1
Introduction
In recent years the neural network has boosted both in the research and in the
application. Modern technologies such as pattern recognition, data mining, nature language processing(NLP), and computer vision massively benefit from the
development of neural network. Among the vast varieties of Neural Networks, the
convolutional neural network(CNN) is the first network which raises the accuracy of image recognition task to a satisfactory level[17]. CNN can extract spatial
features from a local range of the data and raise the abstract level progressively
through its multi-layer structure[18]. Comparing with the early machine learning
techniques such as support vector machine(SVM) and decision trees, CNN has
significantly reduced the artifact work in design and training and promotes the
reusability of a trained network in transfer learning of CNN. For example, the
users can take a trained network to extract the low-abstract-level features and
stack this network with several new layers for classification in high-abstract-level
of their interest[34].
The success of CNN is restricted on Euclidean data such as numbers, 2Dimages, text, and audios. However, non-Euclidean data are ubiquitous in modern
application domains such as 3D-geometry and most of all, graph. Graph, which
is consist of nodes and edges, can represent objects and their interactional relations in a mathematical form. A graph can have sophisticated underlying features behind its structural representation, which is used in social network[31],
in chemistry[25], in biology[21], and in physic controls[26]. As a result, to find
an analogical convolutional method on graph has attracted incremental attention since the success of CNN. However, several challenges prevent the direct
applying of CNN on graph:
2
Min Li
Triangle Inequality not hold. The Triangle Inequality is held in Euclidean
data, which means the shortest path between two points are always a direct
connection between them. In contrary, this feature does not necessarily exist in
a non-Euclidean space such as Manifold. Manifold is a space which is similar to
an Euclidean space locally near a point. One example of Manifold is the surface
of the earth, which was considered as flat in early history. For instance, one man
in Barcelona wants to take a journey to Shanghai. If he is looking at a world
map, he may think that Beijing is located precisely in the east of Barcelona
because the two cities have almost the same latitude on map. If he forgets that
the earth is not flat and sets out to the east along the latitude line, he will suffer
from a longer distance comparing to an alternative path if he heads north at
the first time and crosses the large longitude angle there instead. Therefore, the
shortest path between Barcelona and Shanghai is not the direct connection on
the world map. As a result, the metrics of the similarity between two points
should be different in non-Euclidean space[3].
Transition Invariant not hold. Unlike image and text, on which each point
has a fixed number of neighbors and connections, non-Euclidean data such as
graph has an irregular structure in each node. Consequently, if the center of
the kernel transfers from one center node to another in two convolution steps,
the involved number of nodes and the structure of the calculation changes dramatically. Therefore, the convolution with the kernel, which is the fundamental
operation of CNN, cannot be directly applied on a graph. Besides, the degree of
a node can vary from zero to millions in a large graph, which causes difficulty
in the choice of weight variables of the kernel.
This paper gives a brief overview of development in the domain of graph
convolutional network(GCN) and the basic theoretical idea behind the spectral
approaches. In Section 2, we give a categorization of different approaches as well
as introduce the history of the GNN and related works. In the long Section 3, we
introduces the theoretical foundation of GCN such as the definition of Laplacian
matrix on graph in Subsec. 3.2, the Fourier transformation and decomposition
on graph in Subsec. 3.3, the convolution method on graph in Subsec. 3.4. After
the introduction of the theories, we show three highly correlated approaches
in spectral methods, which are the Non-parametric GCN in Subsec. 3.5, the
ChebyNet in Subsec. 3.6, and the Simplfied ChebyNet in Subsec. 3.7. In Section
4, we show a benchmark of the Simplfied ChebyNet on different datasets. Section
5 concludes this paper and give a dicussion of the future directions.
2
Categorization, History and Related Works
Categorization The graph convolutional network(GCN) is related to and in some
papers categorized as a sub-domain of graph embedding[10][32]. The graph embedding aims to represent the graph nodes in a derived vector space, which
preserves the information on nodes, the structure of the graph, and the edge
between nodes[10].
A Brief Overview of Spectral Graph Convolutional Network
3
Because of the boost in research of the graph embedding and graph neural
network(GNN), different approaches from different aspects have appeared in recent years and makes the categorization complicated and ambiguous. This paper
takes one of the categorization methods which separates the network embedding
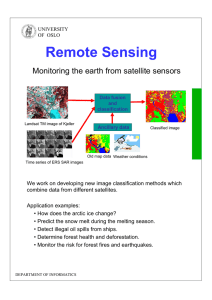
and GNN apart[32]. The separation is due to the rapid development and massive diversity between different GNNs. Fig.1 stats the differences between graph
embedding and GNN. The matrix factorization[1], random walk[22], and LINE
[28] belong to network embedding along while DNGR[30] and SDNE[5] locate
in a shared domain between network embedding and GNN. The GNN contains
four different sub-domains, and this paper focuses only on GCN.
Fig. 1. The categorization method separates graph embedding and GNN apart. The
graph auto-encoder, which contains DNGR and SDNE algorithms, belongs to both
categories due to their neural network alike embedding strategy. The GCN, which is
the focus of this paper, belongs to GNN. Reposted from [32].
Fig. 2. Timeline to show a brief history of graph neural network. In 2005 Macro Gori
finished the first graph neural network(GNN). In 2013 Joan Bruna built the foundament
of spectral methods. In 2016 Defferrard created the ChebyNet and later Kipf and
Welling simplified the ChebyNet. Between the spatial methods, Yujia Li introduced
GGNN algorithm in 2016 and William L. Hamilton published the GraphSage in 2018.
The different approaches of GCN distinguish on their rooted convolution
theory. The approaches which root on the graph spectral theory are widely categorized as spectral methods while the rest approaches as spatial methods[35].
A spectral method usually uses the eigenvalue and the eigenvectors of a graph,
4
Min Li
which should consider the information of all nodes and edges, and is hard to be
parallel and scale to large graphs[32]. A spatial method, in general, performs the
convolution on nodes directly and aggregates the information together, which
allows a good parallelizability and scalability to large graphs[32].
A Brief History A timeline, which is shown in Fig.2, stats several vital developments in the domain of GNN. Macro Gori finished the early work of spatial
method in 2005, which becomes the first GNN in history[9]. A further breakthrough, which is inspired by the huge success of the AlexNet in Computer
vision, took place later in 2013 when Joan Bruna introduced the spectral graph
theory[6] into this domain and built the fundament of all spectral methods[4].
However, the complexity and efficiency of the algorithm in Bruna’s work was still
not satisfactory until Defferrard suggested using Chebychev polynomial as the
kernel function and created the ChebyNet[7]. In 2017 Kipf and Welling simplified
the ChebyNet and finished the most cited paper in the domain of graph convolution[16]. On the other side of the spatial methods, Yujia Li introduced gate
recurrent unit(GRU) from recurrent neural network(RNN) into GNN and established the gated graph sequence neural networks(GGNN)[19]. In 2018 William
L. Hamilton published a simple but well-performing GraphSage algorithm based
on spatial method[11]. There are also many other important developments which
are not mentioned in this paper.
Related Works Several surveys are related to this paper. Bronstein et al.[3] gives
a description in the non-euclidean domain. Wu et al. [32] presents a new taxonomy, which is adapted in this paper, and contributes an extensive introduction as
well as benchmarks in the domain of GNN. Zhou et al.[36] conducts a summary
of all frameworks of the state of art developments[36].
3
3.1
The Spectral Method of GCN
Notations
In this paper, a graph is denoted as G = (V, E) where V = {v1 , ..., vN } is
the set of all N nodes of G and E ⊆ V × V is the set of all edges of G. The
adjacency matrix of graph is denoted as A. This paper only consider the problem
of undirected graph. Most Notations are shown in Table.3.1.
3.2
The Laplacian Matrix
The traditional convolution of CNN cannot be directly applied to non-Euclidean
data, which can be found in Sec. 1. The Laplacian matrix L introduced by Bruna
et al.[4] build the fundament of the convolution with the spectral methods[6].
This section gives a brief description of the Laplacian matrix on graph.
A Brief Overview of Spectral Graph Convolutional Network
5
Notations
Descriptions
Notations
Descriptions
G
Graph
N
Number of nodes
V
Set of all nodes
f,g
Signal function on node
E
Set of all edges
Λ
A diagonal matrix
A
Adjacency Matrix
Â
Aggregated matrix
D
Degree Matrix
D̃
Renormalized matrix
L
Laplacian Matrix
gθ (X)
The kernel/filter
L
Combinatorial Laplacian
dG (i, j)
distance between nodes
Lnorm
Normalized Laplacian
∗
Convolution operator
F
Fourier Transform
◦
Schur product
Table 1. Notations used in this paper.
comb
The finding of Laplacian matrix comes from an analogy with the LaplaceBeltrami operator on Riemannian manifolds[14]. In general, Norman Biggs shows
that the Laplacian Matrix on an undirected graph is equal to the adjacency matrix minus the degree matrix[2]. This Laplacian matrix is also called as Combinatorial Laplacian matrix. The Combinatorial Laplacian matrix is defined as:
Lcomb = A − D
(1)
where Lcomb ∈ RN ×N is the Laplacian matrix of graph G, A ∈ RN ×N is the
adjacency matrix of graph G, D is the degree matrix of graph G. The Equation
1 is proved by [2]
Because this paper only deals with the undirected graph, it exists A(i, j) =
A(j, i) and therefore the adjacency matrix A is symmetry. The value 1 in the
cell A(i, j) represents the existence of the edge between node i and node j while
0 refers to no connection. The degree matrix D is a diagonal matrix with D =
diag(d1 , d2 , ..., dN ) where di represents the degree of node i.
On the diagonal of the Combinatorial Laplacian matrix, the i-th value represents the degree of node i. In other positions L(i, j) out of the diagonal, the
negative 1 stats that the adjacency between node i and node j while 0 stats for no
connection. The following equation summaries the meaning of the Combinatorial
Laplacian matrix:
di
Lcomb (i, j) = −1
0
if i=j,
if i and j are adjacent,
otherwise
as proved by [6].
Chung et al.[6] show that the Combinatorial Laplacian matrix only works
for regular graph and suggest the use of a normalized Laplacian matrix instead,
which is shown in the following equation:
6
Min Li
1
1
1
1
Lnorm = D− 2 Lcomb D− 2 = I − D− 2 AD− 2
norm
N ×N
(2)
N ×N
where L
∈ R
is the normalized Laplacian matrix, A ∈ R
is the
adjacency matrix, D is the degree matrix, I ∈ RN ×N is the identity matrix. The
Equation 2 is proved by [6].
In compare to Combinatorial Laplacian matrix, the diagonal values of the
normalized Laplacian matrix are all ones. The degrees of two adjacent nodes
are relocated to the denominator of Lnorm (i, j), which is shown in the following
equation:
1
1
Lnorm (i, j) = − √di dj
0
if i=j and dv 6= 0
if i and j are adjacent,
otherwise
as proved by [6].
In the rest of this paper, we denote the normalized Laplacian matrix as
Laplacian matrix due to the simplicity. The Laplacian matrix has some essential
features in its eigenvalues and eigenvectors, which can be found in the Sec.3.3.
3.3
Fourier Transformation and Fourier Decomposition on Graph
The spectral graph theory uses Fourier analysis to solve the graph convolution
problem in the spectral domain instead of in the graph domain. The transform
between the spectral and graph domains is based on the Fourier Transformation
on Graph, which is mathematically inspired by the traditional Fourier Transformation[27].
The classical Fourier Transformation can be consider as the inner product of
a function f and a term e−iωt .
Z
−iωt
F (ω) = hf (t), e
i=
f (t)e−iωt dt
(3)
R
where ω is a parameter and t is the time. Some papers write ω = 2πξ instead
while ξ is the frequency[27].
A generalized equation to calculate eigenvectors or eigenfunctions can be
defined as following:
AV = λV
(4)
where A is a certain transformation operator function or transform matrix, V
is a characteristic function or a eigen vector, λ is a scalar which represents the
eigenvalue. The Equation 4 is derived from [23].
If the transformation operator function is the Laplacian operator 4, the
equation turns out to be:
AV = 4e−iωt = ∇2 e−iωt =
∂ 2 −iωt
e
= −ω 2 e−iωt = λV
∂t2
A Brief Overview of Spectral Graph Convolutional Network
7
Therefore, the parameter ω is the eigenvalue of the Laplacian operator function according to the generalized eigenvalue equation.
In analogy, the generalized eigenvalue equation can be applied on Graph with
respect to the Laplacian matrix:
AV = Lul = λl ul = λV
(5)
where L ∈ RN ×N is the Laplacian matrix, ul ∈ RN is the l-th eigenvector of L,
λl ∈ R is the l-th eigenvalue of L.
The eigenvalues and eigenvectors of the Laplacian matrix have three important features: (derived from [24])
Trivial Eigenvector A trivial eigenvector u1 = (1, ..., 1) ∈ RN exists for all
Laplacian matrix, and its corresponding eigenvalue λ1 is 0.
Non-negative Eigenvalues Eigenvalues of the Laplacian matrix are non-negative
and are of real number.
Orthogonal Eigenvectors Eigenvectors of the Laplacian matrix are orthogonal and are of real number.
Graph is a generic data representation form and can represent the data at
each node in the graph, namely graph signal[27]. Inspired by the traditional
Fourier Transform, the Fourier Transform on Graph can also be written as the
inner product between a signal function f and the eigenvector ul :
F (λl ) = hf, ul i =
N
X
f (i)u∗l (i)
i=1
∗
f (λ1 )
u1 (λ1 ) · · · u∗1 (λ1 )
f (λ1 )
.. .. = U ∗ f = U T f
..
fˆ = ... = ...
.
. .
f (λN )
u∗N (λ1 ) · · · u∗N (λ1 )
f (λN )
(6)
where l ∈ RN stats the index of l-th pair of eigenvector and eigenvalue, fˆ ∈
RN is the Fourier transformed signal in spectral domain, f ∈ RN is a certain
signal function on node i in graph domain, u∗l is the conjugate transpose of the
eigenvector ul in the complex space, U ∈ RN ×N stats the matrix of eigenvectors
of Laplacian matrix, which is a matrix of real number. The Equation 6 is derived
from [27].
With the similar derivation, the Inverse Fourier Transform equation on graph
is:
Z
iωt
ˆ
F (t) = hf (ω), e i =
fˆ(ω)eiωt dω
(7)
R
f (1)
u1 (1) · · · uN (1)
fˆ(λ1 )
.. .. = U fˆ
..
f = ... = ...
.
. .
f (N )
u1 (N ) · · · uN (N )
fˆ(λN )
(8)
8
Min Li
where f ∈ RN stats the result of the back transformed signal in graph domain,
and fˆ ∈ RN stats the signal in spectral domain. U ∈ RN ×N stats the matrix of
eigenvectors of Laplacian matrix. The Equation 8 is derived from [27].
The Equation 6 and 8 show that the transform between spectral domain
and graph domain can be achieved by a left scalar product with the eigenvector
matrix or its transposed matrix. In essence, Fourier transform is a change of
bases between the orthonormal bases in graph domain and eigenvector bases in
spectral domain. The Fourier transform on graph makes the signal analysis in
the spectral domain possible, which leads to the Fourier Decomposition
The Equation 7 of the classical Inverse Fourier Transform can be interpreted
as a linear combination of different Fourier bases, which is silimar to the Fourier
Decomposition. In analogy, each pair of ul and λl in the Equation 5 represents a
Fourier base. The eigenvalue represents the importance of the corresponded base,
which can be interpreted as high frequency when the eigenvalue is high and low
frequency when the eigenvalue is low[27]. This notation allows a generalization
between the classical Fourier Decomposition and the Fourier Decomposition on
Graph.
3.4
Convolution on Graph
The traditional convolution theorem in CNN can be concluded as Convolution
operation in time domain is equal to scalar product in frequency domain.
F (f ∗ g) = F (f ) · F (g)
where F is the Fourier transformation, f and g are two signal functions in time
domain. Operator ∗ is the convolution operator and · is the scalar product operator.[15]
In analogy, the convolution theorem can be extended to the graph:
(f ∗ g)(i) = F −1 {F (f ) · F (g)}(i)
=
N
N
X
X
(fˆ(λl ) · ĝ(λl )) ul (λl ) =
fˆ(λl ) ĝ(λl ) ul (λl )
l=1
l=1
If we write the equation in the matrix form:
(f ∗ g)(1)
u1 (1) · · · uN (1)
fˆ(λ1 ) · ĝ(λ1 )
..
..
..
..
..
f ∗g =
= .
.
.
.
.
(f ∗ g)(N )
u1 (N ) · · · uN (N )
fˆ(λN ) · ĝ(λN )
fˆ(λ1 )
ĝ(λ1 )
.. ..
= U . ◦ . = U fˆ ◦ ĝ
ĝ(λN )
fˆ(λN )
A Brief Overview of Spectral Graph Convolutional Network
9
f ∗ g = U fˆ ◦ ĝ = U (U T f ) ◦ (U T g) = U diag fˆ(λ1 ), ..., fˆ(λN ) U T g (9)
where ◦ is the schur product operator. The equation is derived from [4].
The Equation 9 defines the convolution operation on graph, which is the
fundament of the spectral algorithms in Sec. 3.5, 3.6 and 3.7.
3.5
Non-parametric GCN
The Equation 9 defines
the convolution on graph. In 2013 Bruna et al. suggested
using the term diag fˆ(λ1 ), ..., fˆ(λN ) in the Equation 9 as kernel while each
fˆ(λ1 ) is a learnable weight. This algorithm, namely Non-parametric GCN, becomes the first spectral method in the GCN domain. The following equation
shows a forward propagation in a layer:
xk+1 = σ U diag (Fk,1 , ..., Fk,N ) U T xk
(10)
where Fk,i is the i-th learnable weight of the kernel in the k-th layer, σ is a
non-linear activation function, xj is the input of j-th layer. The Equation 10 is
derived from [4].
As an early approach among the spectral methods, some weaknesses are
remarkable in the Non-parametric GCN. The following points of the weaknesses
are summarised by [7]:
High Complexity In each forward propagation step, the calculation of the
term U diag (Fk,1 , ..., Fk,N ) U T has the complexity of O(N 2 ). In addition, the
step of eigenvalue decompositon to get the eigenvector matrix U is also expensive.
Localization Problem One of the keypoints leading to the success of CNN is
its localization in convolution, which allows a feature extraction and aggregation within the size of the kernel. In contrary, the convolution kernel in Nonparametric GCN are N-dimensional, which takes information from all nodes in
graph. The absence of the localization makes it harder to build a multi-layer
model.
Curse of Dimensionality The size of the kernel is N, which is expensive to
process on a large graph.
Because of weaknesses above, this algorithm has never been popular in practice. However, the fundamental idea of its spectral convolution has inspired many
other approaches such as ChebyNet.
3.6
ChebyNet
After analyse the weaknesses of Non-parametric GCN, Defferrard et al. published
the ChebyNet in 2016[7]. The main idea behind the ChebyNet is to limit the
size of the kernel by using the polynomial parametrization.
10
Min Li
The generalized form of the forward propagation, which is filtered by gθ , can
be represented as:
xk+1 = σ (gθ (L) xk ) = σ gθ (U ΛU T ) xk = σ U gθ (Λ) U T xk
where Λ = diag (Fk,1 , ..., Fk,N ) for Non-Parametric GCN, gθ (Λ) is the generalized kernel. [7]
Suppose K is a given setting of the algorithm, which refers to the size of
the kernel. If a polynomial kernel withPa maximalpower of K is applied to
K
approximate the kernel, we get gθ (Λ) = k=0 θk Λk
Suppose that the kernel is centered by node j and the value to be taken in
convolutionPcalculation is from the node i, the calculation step can be written as
K
gθ (Λ)i,j = k=0 θk Lk i,j . According to a lemma from [12], if the shortest path
between two nodes are larger
than k, the polynomial term will be zero, which
follows dG (i, j) > k ⇒ Lk i,j = 0. Therefore, the setting of K can limit the
perception range under the distance of K. In analogy, K is similar to the size of
the kernel in CNN.
A further observation is based on the orthogonality of matrix U :
Lk = (U ΛU T )(U ΛU T ) · · · (U ΛU T ) = (U Λk U T )
Therefore the forward propagation equation with a polynomial kernel can avoid
using the eigenvector matrix U, which can reduce the cost on the eigenvalue
decomposition:
xl+1 = σ U
K
X
k=0
!
k
θk Λ
T
U xk
=σ
K
X
!
k
θk U Λ U
T
k=0
xk
=σ
K
X
!
k
θ k L xl
k=0
The polynomial kernel has solved some problems of the Non-parametric
GCN. Firstly, the polynomial kernel has only K variables of weights, which is
independent on the size N of the input data. Secondly, the expensive eigenvalue
decomposition can be saved. Thirdly, the kernel has a good spatial localization
feature similar to CNN. The only remaining problem is that the complexity of
forward propagation is still O(N 2 ). [7]
To reduce the complexity, Defferrard et al. suggested the employ of Chebyshev polynomial in the approximation:
T0 (x) = 1, T1 (x) = x
Tn+1 (x) = 2xTn (x) − Tn−1 (x)
(11)
Where Ti (x) is the i-th Chebyshev term and x ∈ [−1, 1]. Chebyshev polynomial
is defined recursively.
Before the Chevbychev polynomial replaces the Lk term in the forward propagation equation, the L should be normalized into the range of [−1, 1] with
A Brief Overview of Spectral Graph Convolutional Network
L̃ =
2L
λmax
11
− IN . Finally, the forward propagation equation becomes:
xl+1 = σ
P
K
k=0 θk Tk (L̃)
xl
(12)
T0 (L̃) = 1, T1 (L̃) = L̃
Tk+1 (L̃) = 2xTk (L̃) − Tk−1 (L̃)
Because the Chebyshev term is recursively calculable and a scalar product
with the sparse matrix L̃ has the complexity O(|E|), the using of Chebyshev
polynomial convolution reduces the complexity from O(N 2 ) to O(|E|).[7]
3.7
Simplified ChebyNet
Kipf and Welling simplified the ChebyNet in 2017 and published their simplified
ChebyNet, which is proved to be success in graph learning. This net is sometimes
also called as GCN due to its popularity.
The idea of Kipf & Welling is to limit the size of the kernel K to 1 and stack
multiple convolution layers. Furthermore, the maximal eigenvalue is approximately to be λmax ≈ 2. Thirdly, theparameters θ0 and
θ1 are approximately as
− 21
− 12
is renormalized to raise
one parameter θ. Finally, the term IN − D AD
the numerical stability. The steps of simplification are shown in the following
equations:
gθ (L) =
P
K
k=0 θk Tk (L̃)
− IN x = θ0 x + θ1 (L − IN ) x
1
1
1
1
= θ0 x + θ1 IN − D− 2 AD− 2 − IN x = θ0 x − θ1 D− 2 AD− 2 x
(13)
1
1
1
1
≈ θx − θ D− 2 AD− 2 x = θ IN − D− 2 AD− 2 x
1
1
= θ D̃− 2 AD̃− 2 x
1
1
= ReLU D̃− 2 AD̃− 2 Xl W (l) = ReLU ÂXl W (l)
= θ0 x + θ1
Xl+1
xl
2L
λmax
1
1
Where D̃ is the renormalized degree matrix, Â = D̃− 2 AD̃− 2 is the aggregated
matrix which can be calculated in preprocessing, Xl is the matrix of input data
with different channels in layer l, W (l) is the vector of the learnable weights of
all filters set in the l-th layer. The suggested activation function between layers
is ReLU function. The Equation 13 is proved by [16].
The complexity of the forward propagation in each layer is therefore O(|E|F C),
where |E| is the number of edges on graph, F is the number of filter set in the
layer, C is number of input channels.[16]
The rest configurations are similar to CNN. Kipf and Welling suggest using
softmax with cross entropy behind the last layer for classfication tasks. In traning
process, batch loss, dropout and normal gradient descent are recommended.[16]
12
4
Min Li
Benchmark of Simplified ChebyNet
This benchmark focuses on the accuracy of document classification on three
citation networks, namely Citeseer, Cora and Pubmed, which are the same setup
of node classification stated by Yang et al.[33]. A citation network is composed
of document as node, feature vector of bag-of-words as node feature, citation
link as edge, and exactly one document label as target. We treat the citation
networks as undirected graphs and run various algorithm for classification.
The Fig.3 shows the results of node classification of five common methods
in graph embedding and graph neural network. The tendency of improvement
in accuracy is obvious with the development in this domain. The GraphSage,
which is a spatial method, has already surpassed the spectral method in node
classification task. The three colours represent three different citation network,
on which the training and validation are run. The result of SDNE on Pubmed
network is missing because of the lack of data in the cited paper.[8] [13] [16] [20]
[29]
Fig. 3. The accuracy of node classification. The vertical coordinate stats the accuracy
of node classification of different method on different citation network. The horizontal
coordinate stats the denotation of each method and the year of its release. The three
colours represent three different citation network, on which the training and validation
are run. The tendency of improvement in accuracy is obvious with the development
in this domain. The GraphSage, which is a spatial method, has already surpassed the
spectral method in node classification task. To notice that the result of SDNE on
Pubmed network is missing because of the lack of data in the cited paper. The raw
benchmark results are cited from [8] [13] [16] [20] [29].
A Brief Overview of Spectral Graph Convolutional Network
5
13
Conclusion and Discusssion
In this paper, we provided a brief overview of the spectral methods of GCN. We
first reviewed the history of the domain and introduced a categorization of the
existing methods. The introduction of the spectral methods mainly focused on
the theoretical inference, which follows the logical structure between different
fundaments. The first fundament is the two different Laplacian matrices of the
graph spectral theory, given by their definition and the interpretation on a graph.
The second fundament we showed is the traditional Fourier transformation and
the essence concerning the eigenvalue and eigenvector of the Laplacian operator. In analogy, we presented Fourier transformation on graph between spectral
domain and graph domain, which is closely related to the eigenvalue and eigenvector of the Laplacian matrix. The Inverse Fourier transform equation can be
interpreted as the equation of Fourier decomposition and the eigenvalue as the
frequency of each basis. After defining of Fourier transformation, we derived the
convolution theorem on graph and the equation in the matrix form.
Based on the fundaments, we introduced three approaches of the spectral
methods. The first Non-Parametric GCN method takes the diagonal term in the
convolution equation as their kernel, at the cost of the complexity in calculation
and loss in localization. The ChebyNet uses Chebyshev polynomial to approximate this diagonal term and remarkably reduce the complexity and achieve the
localization of the kernel. The simplified ChebyNet makes some further simplification of the kernel and builds a multilayer model as the CNN. At last, we
presented the benchmark of a node classification task and found the progressive
improvement in the domain. Above all, the accuracy of node classification has
already reached a high level as the AlexNet did in classification of images, which
shows the effectiveness of GCN.
There are some future directions, primitive ideas and possible application
which I have noticed:
The future of spatial methods is more brilliant Although the first breakthrough of GCN comes from the spectral approaches, the tendency of development goes towards the spatial methods. One reason for the success of simplified
ChebyNet is the limitation of perceptive field of a centered kernel. Therefore, the
simplified ChebyNet can be regarded as an approximation of a spatial method in
the spectral domain. In the benchmark section, we have shown that the GraphSage method, which is a spatial method, has surpassed the accuracy of simplified
ChebyNet. Furthermore, the spectral method is generally hard to run in parallel
because the Fourier transformation takes the whole graph in the calculation.
The spectral methods can still have its application scenario in some cases, but
the main focus of research has shifted to the spatial methods in last year.
Different Graphs All introduced methods of this paper is for general undirected graphs. In the world of graphs, there are still more special graphs such
as heterogenous graph, directed graph, etc. Each special graph has its special
structure and can have respective optimization methods.
The variable size of the perceptive field Each node in the graph has its
special betweenness, which shows the importance that one node is for a network.
14
Min Li
In the aspect of information flow, a node with high betweenness has a higher
possibility to touch information from all directions while a node with low betweenness plays a localized role. Therefore, we can enlarge the receptive field
of the high betweenness nodes to convolute more information together for ambiguous but extensive knowledge, while reducing the receptive field of the low
betweenness nodes for localized but explicit knowledge.
Deep-Dream on graph Deep-Dream, a network to creates a new image from
learned knowledge, has shown its success in Computer Vision. In analogy, the
graph convolutional network should also have the ability to create a graph or
network based on its learned knowledge. This technique could have some application field, such as the design of an isomer of medicine and the creation of a
new agriculture product with gene editing.
Learning algorithm for learning algorithms The existing learning algorithms such as CNN, RNN and GCN are graph in essence. It could be possible
to train a network to generate a suitable learning algorithm according to the
given problem and the training and validation data set.
References
1. Ahmed, A., Shervashidze, N., Narayanamurthy, S., Josifovski, V., Smola, A.J.:
Distributed large-scale natural graph factorization pp. 37–48 (2013)
2. Biggs, N., Biggs, N., Norman, B., Press, C.U.: Algebraic Graph Theory. Cambridge
Mathematical Library, Cambridge University Press (1993)
3. Bronstein, M.M., Bruna, J., LeCun, Y., Szlam, A., Vandergheynst, P.: Geometric
deep learning: going beyond euclidean data. CoRR abs/1611.08097 (2016)
4. Bruna, J., Zaremba, W., Szlam, A., Lecun, Y.: Spectral networks and locally connected networks on graphs (12 2013)
5. Cao, S., Lu, W., Xu, Q.: Deep neural networks for learning graph representations
(2016)
6. Chung, F.R.K.: Spectral Graph Theory. American Mathematical Society (1997)
7. Defferrard, M., Bresson, X., Vandergheynst, P.: Convolutional neural networks on
graphs with fast localized spectral filtering. CoRR abs/1606.09375 (2016)
8. Gao, H., Huang, H.: Deep attributed network embedding pp. 3364–3370
9. Gori, M., Monfardini, G., Scarselli, F.: A new model for learning in graph domains
2, 729–734 vol. 2 (July 2005)
10. Goyal, P., Ferrara, E.: Graph embedding techniques, applications, and performance: A survey. CoRR abs/1705.02801 (2017)
11. Hamilton, W.L., Ying, R., Leskovec, J.: Inductive representation learning on large
graphs. CoRR abs/1706.02216 (2017)
12. Hammond, D., Vandergheynst, P., Gribonval, R.: Wavelets on graphs via spectral
graph theory. Applied and Computational Harmonic Analysis 30, 129–150 (12
2009)
13. Huang, W., Zhang, T., Rong, Y., Huang, J.: Adaptive sampling towards fast graph
representation learning. CoRR abs/1809.05343 (2018)
14. Jakobson, D., Miller, S.D., Rivin, I., Rudnick, Z.: Eigenvalue Spacings for Regular
Graphs. Springer New York (1999)
15. Katznelson, Y.: An introduction to harmonic analysis (1976)
A Brief Overview of Spectral Graph Convolutional Network
15
16. Kipf, T.N., Welling, M.: Semi-supervised classification with graph convolutional
networks. CoRR abs/1609.02907 (2017)
17. Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks pp. 1097–1105 (2012)
18. LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–44 (05 2015)
19. Li, Y., Tarlow, D., Brockschmidt, M., Zemel, R.S.: Gated graph sequence neural
networks. CoRR abs/1511.05493 (2016)
20. Ma, Z., Li, M., Wang, Y.: PAN: path integral based convolution for deep graph neural networks. CoRR abs/1904.10996 (2019), http://arxiv.org/abs/1904.10996
21. Mason, O., Verwoerd, M.: Graph theory and networks in biology. IET Systems
Biology 1(2), 89–119 (March 2007)
22. Perozzi, B., Al-Rfou, R., Skiena, S.: Deepwalk: Online learning of social representations. CoRR abs/1403.6652 (2014)
23. Putinar, M.: Generalized eigenfunction expansions and spectral decompositions.
Banach Center Publications 38 (1997)
24. Rajaraman, A., Ullman, J.D.: Mining of massive datasets. Cambridge University
Press, Cambridge (2012)
25. Randic, M.: Nenad trinajstic - pioneer of chemical graph theory. Croatica Chemica
Acta 77, 1–15 (05 2004)
26. Sanchez-Gonzalez, A., Heess, N., Springenberg, J.T., Merel, J., Riedmiller, M.A.,
Hadsell, R., Battaglia, P.: Graph networks as learnable physics engines for inference
and control. CoRR abs/1806.01242 (2018)
27. Shuman, D.I., Narang, S.K., Frossard, P., Ortega, A., Vandergheynst, P.: Signal
processing on graphs: Extending high-dimensional data analysis to networks and
other irregular data domains. CoRR abs/1211.0053 (2012)
28. Tang, J., Qu, M., Wang, M., Zhang, M., Yan, J., Mei, Q.: LINE: large-scale information network embedding. CoRR abs/1503.03578 (2015)
29. Tu, C., Zeng, X., Wang, H., Zhang, Z., Liu, Z., Sun, M., Zhang, B., Lin, L.: A
unified framework for community detection and network representation learning.
IEEE Transactions on Knowledge and Data Engineering 31(6), 1051–1065 (2019)
30. Wang, D., Cui, P., Zhu, W.: Structural deep network embedding pp. 1225–1234
(2016)
31. Wasserman, S., Faust, K.: Social network analysis: Methods and applications,
vol. 8. Cambridge university press (1994)
32. Wu, Z., Pan, S., Chen, F., Long, G., Zhang, C., Yu, P.S.: A comprehensive survey
on graph neural networks. CoRR abs/1901.00596 (2019)
33. Yang, Z., Cohen, W.W., Salakhutdinov, R.: Revisiting semi-supervised learning
with graph embeddings. CoRR abs/1603.08861 (2016)
34. Yosinski, J., Clune, J., Bengio, Y., Lipson, H.: How transferable are features in
deep neural networks? CoRR abs/1411.1792 (2014)
35. Zhang, Z., Cui, P., Zhu, W.: Deep learning on graphs: A survey. CoRR
abs/1812.04202 (2018)
36. Zhou, J., Cui, G., Zhang, Z., Yang, C., Liu, Z., Sun, M.: Graph neural networks:
A review of methods and applications. CoRR abs/1812.08434 (2018)