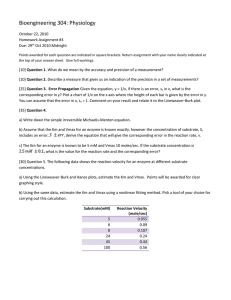
MODELLING, SIMULATION AND OPTIMIZATION OF BIOPROCESSES BT401 Dr.R.Satish Babu CONTENTS • Introduction • Modelling Principles & Fundamentals of Modelling • Modeling approaches • Neural network modeling • Simulation using Berkely Madonna Program • Optimization What is “Model” Model • Three-dimensional representation of a person or thing or of a proposed structure, typically on a smaller scale than the original • synonyms:replicareplica, copyreplica, copy, re presentationreplica, copy, representation, mo ck-upreplica, copy, representation, mock-up, d ummyreplica, copy, representation, mock-up, dummy, imitationreplica, copy, representation , mock-up, dummy, imitation, doublereplica, c opy, representation, mock-up, dummy, imitati Model? • A thing used as an example to follow or imitate • synonyms:prototypeprototype, stereotypepro totype, stereotype, archetype, • versionversion, style. What is a Scientific Model? • A scientific model is a representation of a particular phenomenon in the world using something else to represent it, making it easier to understand • A scientific model could be a diagram or picture, a physical model like an aircraft model, a computer program, or set of complex mathematics that describes a situation. Model? • Graphical / Physical (Visual Models), mathematical (symbolic)representation or simplified version of a concept • Visual models are things like flowcharts, pictures, and diagrams that help us educate each other Use? • The goal is to make the particular thing you're modeling easier to understand. • When we do that, we're able to use it to predict what will happen in the future. • For example, predicting what will happen as our climate changes would be easy if we could make a fully accurate model of the atmosphere. • A representation of a system that allows for investigation of the properties of the system and, in some cases, prediction of future. • Scientific models are often mathematical models, where you use math to describe a particular phenomenon. • For example, M-M model Use of models in Bioprocess? • Use of Models for Understanding, Design and Optimization of Bioreactors Use of models in Bioprocess? Mechanistic models Fig.3 Classification of models as mechanistic and nonmechanistic Different perspectives for cell population kinetic representation (Adapted from Bailley and Ollis,1986) Integrative dynamic model Integrative simultaneous saccharification and fermentation (SSF) model. The SSF model is a combination of (1) enzyme hydrolysis model describing the kinetics of enzymatic hydrolysis of lignocellulosic biomass and (2) integrative dynamic model describing cell growth and fermentation kinetics (adapted from Unrean et al., submitted) Why Bioprocess is so complex? Fig.2 Certain important parameters, phenomena, and interactions which determine cell population kinetics (Adapted from Bailley and Ollis,1986) Nonlinear Models Approaches for model development Empirical Approach • Measure productivity for all combinations of reactor operating conditions, and make correlations. • - Advantage: Little thought is necessary • - Disadvantage: Many experiments are required. Modelling Approach • Establish a model and design experiments to determine the model parameters. • Compare the model behaviour with the experimental measurements. • Use the model for rational design, control and optimization. • - Advantage: Fewer experiments are required, and greater understanding is obtained. • - Disadvantage: Some strenuous thinking may be necessary. Statistical modeling • In statistical modeling, regression analysis is a set of statistical processes for estimating the relationships between a dependent variable and one or more independent variables. Objectives/De pendent Independent Enzyme activity Product Yield % degradation Time Temp pH RPM D.O Limiting substrate & product concentrations Concentration of inhibitor etc Statistical models • Enzyme Activity (Y1) =89.19+4.07A+ 9.37B + 0.55C + 0.82D+ 0.68E+0.034AB-1.12AC • Yield (Y2)=7.90+0.52A + 0.16B + 0.018C + 0.27D + 0.14E + 0.072AB + 0.041AC + 0.047AD • Where A,B,C and D are independent variables Concepts of mass balance models General Modelling Procedure the following stages in the modelling procedure • The first stage involves the proper definition of the problem and hence the goals and objectives of the study. • All the relevant theory must then be assessed in combination with any practical experience with the process. • (ii)The available theory must then be formulated in mathematical terms. • Most bioreactor operations involve quite a large number of variables (cell, substrate and product concentrations, rates of growth, consumption and production) and many of these vary as functions of time (batch, fed-batch operation). • For these reasons the resulting mathematical relationships often consist of quite large sets of differential equations. • (iii) Having developed a model, the model equations must then be solved. • Mathematical models of biological systems are usually quite complex and highly non-linear. • Numerical methods of solution must therefore be employed, with the method preferred in this text being that of digital simulation. General Aspects of the Modelling Approach (Mass & Energy balances) • An essential stage in the development of any model, is the formulation of the appropriate mass and energy balance equations. • It is required to add appropriate kinetic equations for rates of cell growth, substrate consumption and product formation, equations • The combination of these relationships provides a basis for the quantitative description of the process and comprises the basic mathematical model. • The resulting model can range from a very simple case of relatively few equations to models of very great complexity. • Simple models are often very useful, since they can be used to determine the numerical values for many important process parameters. • For example, a model based on a simple Monod kinetics can be used to determine basic parameter values such as the specific growth rate (μ), saturation constant (Ks), biomass yield coefficient (YX/s) and maintenance coefficient (m). • A basic use of a process model is to analyze experimental data and to use this to characterize the process, by assigning numerical values to the important process variables. • The model can then also be solved with appropriate numerical data values and the model predictions compared with actual practical results. • This procedure is known as simulation and may be used to confirm that the model and the appropriate parameter values are "correct". Simulations, however, can also be used in a predictive manner to test probable behavior under varying conditions. • This leads on to the use of models for process optimization and their use in advanced control strategies. The application of a combined modelling and simulation approach leads to the following advantages: • Modelling improves understanding. • Forced to consider the complex cause-and-effect sequences of the process • The comparison of a model prediction with actual behaviour usually leads to an increased understanding of the process. • The results of a simulation can also often suggest reasons as to why certain observed, and apparently inexplicable, phenomena occur in practice. • Models help in experimental design. It is important that experiments be designed in such a way that the model can be properly tested. • Mathematical models can also be used for the design of relatively sophisticated control algorithms. • Both mathematical and knowledge based models can be used in designing and optimizing new processes. • Models may be used in training and education. • Many important aspects of bioreactor operation can be simulated by the use of very simple models. • These include such concepts as linear growth, double substrate limitation, changeover from batch to fed-batch operation dynamics, fedbatch feeding strategies, aeration dynamics, measurement probe dynamics, cell retention systems, microbial interactions, biofilm diffusion and bioreactor control. • Models may be used for process optimization. Optimization usually involves considering the influence of two or more variables, often one directly related to the product. Product Balance V dP1 dt = rx V Berkely Madonna Simulation Program for Batch reactor • • • • • • • • • • METHOD RK4 STARTTIME = 0 STOPTIME=10 DT = 0.02 UM = 0.3 KS = 0.1 Y = 0.8 INIT X = 0.01 INIT S = 10 INIT P = 0 • • • • • • • X' = RX ; S' = RS ; P' = RP ; RX = U*X RS=-RX/Y RP=(K1+K2*U)*X U=UM*S/(KS+S) Batch Reactor UM = 0.3 KS = 0.1 X0 = 0.1 S0 = 5 P0 = 0.25 Y1=0.8 Y2=0.7 INIT X = 0.1 INIT S = 5 INIT P = 0.25 X'=RX ; S'=RS ; P'=RP ; RX = U*X U=UM*S/(KS CHEMOSTAT Fed Batch UM = 0.3 KS = 0.1 X0 = 0.1 S0 = 5 P0 = 0.25 D=0.25 Y1=0.8 Y2=0.7 UM = 0.3 KS = 0.1 Y1 = 0.8 Y2=0.7 X0 = 0.1 S0 = 10 P0 = 0.0 F=25 INIT X = 0.1 INIT S = 5 INIT P = 0.25 INIT V=1 INIT VX=V*X0 INIT VS=V*S0 INIT VP=V*P0 X'=D*(X0-X) +RX ; S'=D*(S0-S) +RS ; P'=D*(P0-P) V'=F ; VX'=RX*V ; Assume: Sterile feed VS'=F*S0-RS*V ; VP'=RP*V; X = VX/V S=VS/V P=VP/V U=UM*S/(KS+S) RX=U*X RS=RX/Y1 RP=Y2*U*X D=F/V Infectious disease modelling SIR Model Framework • SIR epidemic model S I R • Large population • Single initial case • Rest of the population is fully susceptible Examples: – – – – Influenza pandemics SARS Ebola Covid-19 The equations Equations: Initial conditions: Parameters: Assumptions: ▪ Homogeneous mixing ▪ Constant recovering rate SIR output: the epidemic curve I R Susceptible Removed Proportion of population S Infectious Time Full dynamics Enzyme Kinetics - Inhibition Types of Inhibition • • • • Competitive Inhibition Noncompetitive Inhibition Uncompetitive Inhibition Irreversible Inhibition Competitive Inhibition In competitive inhibition, the inhibitor competes with the substrate for the same binding site Competitive Inhibition - Reaction Mechanism In competitive inhibition, the inhibitor binds only to the free enzyme, not to the ES complex General Michaelis-Menten Equation This form of the Michaelis-Menten equation can be used to understand how each type of inhibitor affects the reaction rate curve In competitive inhibition, only the apparent Km is affected (Km,app> Km), The Vmax remains unchanged by the presence of the inhibitor. Competitive inhibitors alter the apparent Km, not the Vmax Vmax,app = Vmax Km,app > Km The Lineweaver-Burk plot is diagnostic for competitive inhibition Relating the Michaelis-Menten equation, the v vs. [S] plot, and the physical picture of competitive inhibition Inhibitor competes with substrate, decreasing its apparent affinity: Km,app > Km Formation FormationofofEI EI complex complex shifts shiftsreaction reaction to >>KKm to the the left: left:KKm,app m,app m Km,app > Km Vmax,app = Vmax Example - Competitive Inhibition Sulfanilamide is a competitive inhibitor of p-aminobenzoic acid. Sulfanilamides (also known as sulfa drugs, discovered in the 1930s) were the first effective systemic antibacterial agents. Because we do not make folic acid, sulfanilamides do not affect human cells. Practical case: Methanol poisoning A wealthy visitor is taken to the emergency room, where he is diagnosed with methanol poisoning. You are contacted by a 3rd year NIT student and asked what to do? How would you suggest treating this patient? Methanol (CH3OH) is metabolized to formaldehyde and formic acid by alcohol dehydrogenase. You advisethe third year student to get the patient very drunk. Since ethanol (CH3CH2OH) competes with methanol for the same binding site on alcohol dehydrogenase, it slows the metabolism of methanol, allowing the toxic metabolites to be disposed of before they build up to dangerous levels. By the way, the patient was very grateful and decided to leave all their worldly possessions to the hospital. Unfortunately, after being released from the hospital, he went to the casinos and lost everything he had. Noncompetitive Inhibition the inhibitor does not interfere with substrate binding (and vice versa) Noncompetitive Inhibition Reaction Mechanism In noncompetitive inhibition, the inhibitor binds enzyme irregardless of whether the substrate is bound Noncompetitive inhibitors decrease the Vmax,app, but don’t affect the Km Vmax,app < Vmax Km,app = Km Why does Km,app = Km for noncompetitive inhibition? The inhibitor binds equally well to free enzyme and the ES complex, so it doesn’t alter apparent affinity of the enzyme for the substrate The Lineweaver-Burk plot is diagnostic for noncompetitive inhibition Relating the Michaelis-Menten equation, the v vs. [S] plot, and the physical picture of noncompetitive inhibition Inhibitor doesn’t interfere with substrate binding, Km,app = Km Even at high substrate levels, Formation inhibitor of EI still binds, [E]t < reaction [ES] complex shifts Vmax,app < Vmax to the left: Km,app > Km Vmax,app Km,app > K< mVmax Vmax,app ==V K Kmax m,app m Noncompetitive inhibitors decrease the apparent Vmax, but do not alter the Km of the reaction Example of noncompetitive inhibition: fructose 1,6-bisphosphatase inhibition by AMP Fructose 1,6-bisphosphatase is a key regulatory enzyme in the gluconeogenesis pathway. High amounts of AMP signal that ATP levels are low and gluconeogenesis should be shut down while glycolysis is turned on. High AMP levels inhibit fructose 1,6-bisphosphatase (shutting down gluconeogenesis) and activate phosphofructokinase (turning on glycolysis). Regulation of fructose 1,6-bisphosphatase and phosphofructokinase by AMP prevents a futile cycle in which glucose is simultaneously synthesized and broken down. Uncompetitive Inhibition In uncompetitive inhibition, the inhibitor binds only to the ES complex Uncompetitive Inhibition Reaction Mechanism In uncompetitive inhibition, the inhibitor binds only to the ES complex, it does not bind to the free enzyme Uncompetitive inhibitors decrease both the Vmax,app and the Km,app Vmax,app < Vmax Km,app < Km Notice that at low substrate concentrations, uncompetitive inhibitors have little effect on the reaction rate because the lower Km,app of the enzyme offsets the decreased Vmax,app Uncompetitive inhibitors decrease both the Vmax,app and the Km,app of the enzyme Notice that uncompetitive inhibitors don’t bind to the free enzyme, so there is no EI complex in the reaction mechanism The Lineweaver-Burk plot is diagnostic for uncompetitive inhibition Relating the Michaelis-Menten equation, the v vs. [S] plot, and the physical picture of uncompetitive inhibition Inhibitor increases the amount of enzyme bound to substrate Km,app < Km Even at high substrate Formation of EI levels, inhibitor binds, complex shifts reaction [E]t < [ES] to the left: KVm,app >< V Km max,app max Vmax,app < Vmax Km,app< Km Uncompetitive inhibitors decrease the apparent Km of the enzyme and decrease the Vmax of the reaction