Lab 6: Dimensionality Reduction
In this lab, we will explore dimensionality reduction, which can be divided into two components: Feature
Selection and Feature Extraction.
Feature selection is a process by which we automatically search for the best subset of attributes in our dataset.
The notion of “best” is relative to the problem we are trying to solve, but typically means the highest
performance score or the lowest error.
Feature extraction reduces the data in a high dimensional space to a lower dimension space.
Three key benefits of performing dimensionality reduction on the data are:
Reduces Overfitting: Less redundant data means less opportunity to make decisions based on noise
Improves Accuracy: Less misleading data means modelling accuracy improves
Reduces Training Time: Less data means that algorithms train faster
Prepare and Explore Data
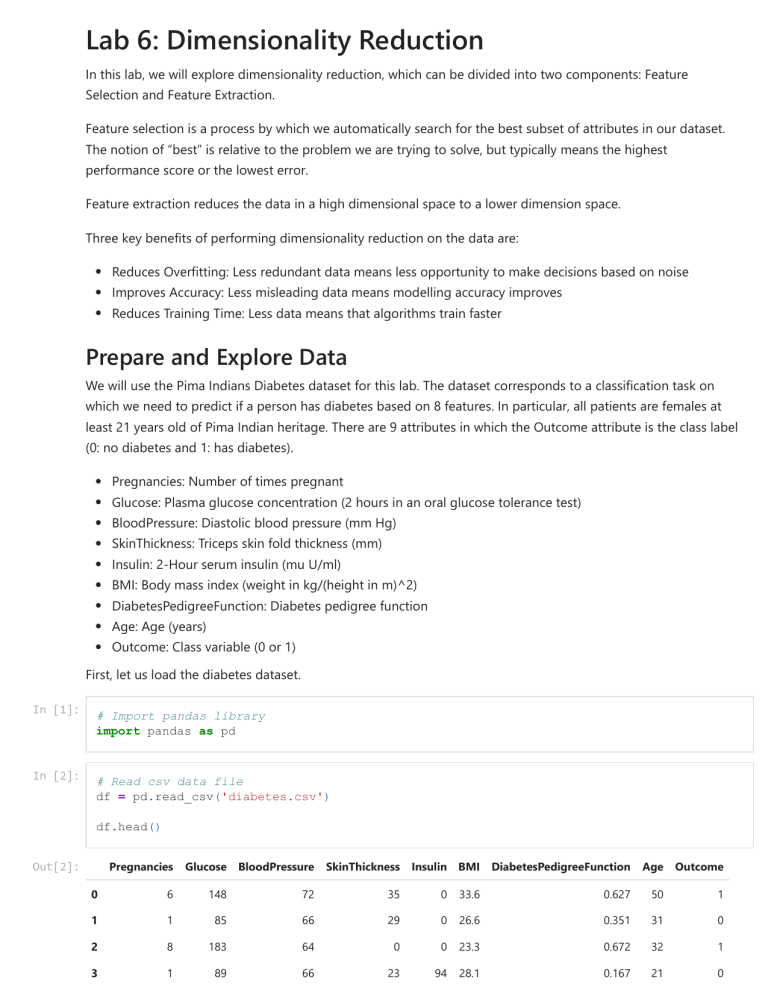
We will use the Pima Indians Diabetes dataset for this lab. The dataset corresponds to a classification task on
which we need to predict if a person has diabetes based on 8 features. In particular, all patients are females at
least 21 years old of Pima Indian heritage. There are 9 attributes in which the Outcome attribute is the class label
(0: no diabetes and 1: has diabetes).
Pregnancies: Number of times pregnant
Glucose: Plasma glucose concentration (2 hours in an oral glucose tolerance test)
BloodPressure: Diastolic blood pressure (mm Hg)
SkinThickness: Triceps skin fold thickness (mm)
Insulin: 2-Hour serum insulin (mu U/ml)
BMI: Body mass index (weight in kg/(height in m)^2)
DiabetesPedigreeFunction: Diabetes pedigree function
Age: Age (years)
Outcome: Class variable (0 or 1)
First, let us load the diabetes dataset.
In [1]:
In [2]:
# Import pandas library
import pandas as pd
# Read csv data file
df = pd.read_csv('diabetes.csv')
df.head()
Pregnancies
Glucose
BloodPressure
SkinThickness
Insulin
BMI
DiabetesPedigreeFunction
Age
Outcome
0
6
148
72
35
0
33.6
0.627
50
1
1
1
85
66
29
0
26.6
0.351
31
0
2
8
183
64
0
0
23.3
0.672
32
1
3
1
89
66
23
94
28.1
0.167
21
0
Out[2]:
Pregnancies
Glucose
BloodPressure
SkinThickness
Insulin
BMI
DiabetesPedigreeFunction
Age
Outcome
0
137
40
35
168
43.1
2.288
33
1
4
In [3]:
# Display the summary of data
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
#
Column
Non-Null Count
--- ------------------0
Pregnancies
768 non-null
1
Glucose
768 non-null
2
BloodPressure
768 non-null
3
SkinThickness
768 non-null
4
Insulin
768 non-null
5
BMI
768 non-null
6
DiabetesPedigreeFunction 768 non-null
7
Age
768 non-null
8
Outcome
768 non-null
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
Dtype
----int64
int64
int64
int64
int64
float64
float64
int64
int64
We first examine the features to gain a better understanding of the data types we are working with. From the
summary, we observe that all 9 attributes are stored as continuous variables. Since this is a classification task,
Outcome should be a categorical variable but we will not convert Outcome yet to a categorical variable (we will
do the conversion later).
In [4]:
Out[4]:
# Find out the number of instances and attributes
df.shape
(768, 9)
There are 768 instances and 9 attributes in the dataset.
In [4]:
# Indicate the target column
target = df['Outcome']
# Indicate the columns that will serve as features
features = df.drop('Outcome', axis = 1)
In [5]:
# Split data into train and test sets
# Import train_test_split function
from sklearn.model_selection import train_test_split
# Split the dataset into training and test sets
x_train, x_test, y_train, y_test = train_test_split(features, target, \
test_size = 0.2, random_state = 0)
We will select and extract features from the training set.
Feature Selection
We will implement two approaches to perform feature selection.
Filter approach
Wrapper approach
Filter Approach
The filter approach will take only the subset of the relevant features based on certain statistical measure and
does not involve any machine learning algorithm. There are many statistical measures that can be used for filterbased feature selection. As such, the choice of statistical measures is highly dependent upon the feature and
target variable data types. The data type can either be continuous/numerical or categorical.
We will discuss four filter approaches:
1. Variance threshold
2. Correlation coefficient
3. Chi-squared
4. Information gain
Variance Threshold
Removing features with low variance. Motivated by the idea that low variance features contain less information.
Calculate variance of each feature. Then, drop features with variance below some threshold. Important to make
sure features have the same scale.
In order to use this filter, we will first have to transform the features to have the same scale. In previous lab, we
have experimented with data standardization for feature scaling. We will experiment with another feature scaling
method known as data normalization.
Normalizing in scikit-learn refers to rescaling each observation (row) to have a length of 1. After applying
normalization, scaler transforms features into an array data structure (not in the form of the data frame
anymore).
In [6]:
# Import normalizer module
from sklearn import preprocessing
# Create the Scaler object
scaler = preprocessing.Normalizer()
# Fit the data on the Scaler object
scaled_features = scaler.fit_transform(x_train)
# View the first 5 rows of scaled_features array
scaled_features[0:5]
Out[6]:
array([[3.14609065e-02,
5.66296317e-01,
[3.28427192e-02,
0.00000000e+00,
[0.00000000e+00,
9.59672638e-01,
[5.35681315e-03,
7.23169775e-01,
[6.17065882e-02,
0.00000000e+00,
6.74162282e-01,
1.58203415e-01,
7.96435940e-01,
2.31541170e-01,
2.32861743e-01,
7.38101161e-02,
5.83892633e-01,
1.34991691e-01,
9.25598823e-01,
2.31399706e-01,
3.50564386e-01,
3.11013533e-03,
4.92640787e-01,
3.63733115e-03,
1.27015496e-01,
6.02617965e-04,
2.99981536e-01,
4.46222535e-03,
0.00000000e+00,
1.41153821e-03,
1.30338041e-01,
2.42698421e-01],
1.88845635e-01,
1.80634955e-01],
4.65723486e-02,
3.24595157e-02],
1.12493076e-01,
1.23206702e-01],
0.00000000e+00,
2.93106294e-01]])
VarianceThreshold is a simple baseline approach to feature selection. It removes all features with variance that
do not meet some threshold. We have to set this threshold parameter. If we are not sure what value of threshold
to set, we can first examine the variance of each column. Variance usually only makes sense for continuous
variables.
In [7]:
names = x_train.columns
# After normalization, scaled_features is transformed into an array so we need to convert
scaled_features_df = pd.DataFrame(scaled_features, columns = names)
# Compute the variance of each column
scaled_features_df.var()
Out[7]:
Pregnancies
Glucose
BloodPressure
SkinThickness
Insulin
BMI
DiabetesPedigreeFunction
Age
dtype: float64
0.000457
0.026139
0.023945
0.008652
0.116097
0.003973
0.000004
0.006816
From the column variance observed, if we set the variance threshold too high, no features would be returned.
Let us set the variance threshold = 0.1.
In [8]:
# Import VarianceThreshold module
from sklearn.feature_selection import VarianceThreshold
# Create VarianceThreshold object with a variance threshold of 0.1
thresholder = VarianceThreshold(threshold = 0.1)
# Conduct variance thresholding - fit_transform() takes in an array
features_high_variance = thresholder.fit_transform(scaled_features)
# Use the get_support() function to identify the feature(s) above the variance threshold
thresholder.get_support(indices = True)
Out[8]:
array([4], dtype=int64)
[0] Pregnancies
[1] Glucose
[2] BloodPressure
[3] SkinThickness
[4] Insulin
[5] BMI
[6] DiabetesPedigreeFunction
[7] Age
Array index starts from [0]. Index [4] indicates column 5: Insulin. Only one feature is returned.
When to use variance threshold filter
As variance is a statistical measure for continuous variables, VarianceThreshold makes more sense to be used
when features are continuous variables. When applied on categorical variables, the interpretation of variance
may not make a lot of sense.
Correlation Coefficient
One way to select features that are good predictors of the target is to identify features that are highly correlated
with the target. We can compute Pearson correlation between each feature and the target. Sklearn's
feature_selection module does not provide a filter based on correlation coefficient but we can generate a
correlation matrix to select good features.
The correlation coefficient has values between -1 to 1.
A value closer to 0 implies weaker correlation (exact 0 implying no correlation)
A value closer to 1 implies stronger positive correlation
A value closer to -1 implies stronger negative correlation
Correlation coefficient only works for continuous variables. This filter approach is suitable when both features
and target are continuous variables. This is the reason why we did not convert Outcome from integer to string
earlier so we can compute the correlation between each feature to the target. We usually do not use the
correlation coefficient filter when our target is a categorical variable.
In [9]:
# Place the x_train and y_train data frames side by side
x = pd.concat([x_train, y_train], axis = 1)
# Generate correlation matrix
cor = x.corr()
# Print correlation matrix
cor
Pregnancies
Glucose
BloodPressure
SkinThickness
Insulin
BMI
Pregnancies
1.000000
0.127642
0.141417
-0.084695
-0.080762
0.003036
Glucose
0.127642
1.000000
0.147744
0.057374
0.326961
0.234836
BloodPressure
0.141417
0.147744
1.000000
0.214119
0.081911
0.264209
SkinThickness
-0.084695
0.057374
0.214119
1.000000
0.426754
0.405780
Insulin
-0.080762
0.326961
0.081911
0.426754
1.000000
0.192086
BMI
0.003036
0.234836
0.264209
0.405780
0.192086
1.000000
DiabetesPedigreeFunction
-0.047203
0.118450
0.052385
0.194594
0.189132
0.145073
Age
0.539582
0.278075
0.229556
-0.156347
-0.063597
0.009823
Outcome
0.193991
0.459278
0.057662
0.088248
0.119886
0.303850
Out[9]:
In [10]:
# Generating the correlation heatmap is optional
# The heatmap is just a visualization of the correlation matrix
# Import seaborn package to generate heatmap
import seaborn as sns
# Import pyplot to control the size of the plot
import matplotlib.pyplot as plt
# Set plot size
plt.figure(figsize=(12,10))
# Generate the heatmap
sns.heatmap(
cor,
vmin = -1, vmax = 1, center = 0,
cmap = sns.diverging_palette(20, 220, n=200),
square = True,
annot = True
)
Out[10]:
<AxesSubplot:>
DiabetesPedigree
If you are interested to learn more about the heatmap parameter, check out:
https://seaborn.pydata.org/generated/seaborn.heatmap.html. The darker the color of the cell, the higher the
correlation.
In [11]:
# Select features above a correlation threshold to target
# Correlation with target
# Apply abs() to get the absolute value so no need to deal with negative correlations
cor_target = abs(cor['Outcome'])
# Selecting highly correlated features
# Say we set the correlation threshold to 0.2
relevant_features = cor_target[cor_target > 0.2]
relevant_features
Out[11]:
Glucose
0.459278
BMI
0.303850
Age
0.238986
Outcome
1.000000
Name: Outcome, dtype: float64
As we can observe, only the features [Glucose, BMI, Age] have correlation above the threshold with the target
variable Outcome. Hence, we will drop all other features apart from these. We can even further reduce the
selected features in the subset. Ideally, we want to keep features that are independent variables that are
uncorrelated with each other. If these variables in the selected feature subset are correlated with each other,
then we need to keep only one of them and drop the rest. This can be done by visually checking the correlation
matrix above.
When to use correlation coefficient filter
When the data type of our feature to be tested and the target variable are both continuous.
Chi-squared
Chi-squared is a goodness of fit statistic. We can use sklearn's univariate feature selection to select the best
features based on univariate statistical tests. We will use SelectKBest function to remove all but the highest
scoring features. The k parameter indicates the number of top features to select.
In [5]:
Out[5]:
In [5]:
Out[5]:
In [8]:
# Currently the target is integer data type
y_train.dtype
dtype('int64')
# Convert integer to string
y_train.astype(str)
603
118
247
157
468
1
0
0
0
1
..
763
0
192
1
629
0
559
0
684
0
Name: Outcome, Length: 614, dtype: object
# Import SelectKBest and chi2 modules
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
# Create a selector
# Setting k = 3 means we want the top 3 features
selector = SelectKBest(chi2, k = 3)
# Select top 3 features based on the training set
x_new = selector.fit_transform(x_train, y_train)
selector.get_support(indices=True)
Out[8]:
array([1, 4, 7], dtype=int64)
[0] Pregnancies
[1] Glucose
[2] BloodPressure
[3] SkinThickness
[4] Insulin
[5] BMI
[6] DiabetesPedigreeFunction
[7] Age
The three best selected features are index[1], index[4] and index[7]: Glucose, Insulin and Age.
When to use Chi-squared
When the data type of our feature to be tested and the target variable are both categorical.
Information Gain
The information gain filter can be implemented using mutual_info_classif.
In [107…
# Import SelectKBest and mutual_info_classif modules
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_classif
# Create a selector
# Setting k = 3 means we want the top 3 features
selector = SelectKBest(mutual_info_classif, k = 3)
# Select top 3 features based on the training set
x_new = selector.fit_transform(x_train, y_train)
selector.get_support(indices=True)
Out[107…
array([1, 5, 7], dtype=int64)
[0] Pregnancies
[1] Glucose
[2] BloodPressure
[3] SkinThickness
[4] Insulin
[5] BMI
[6] DiabetesPedigreeFunction
[7] Age
The two best selected features are index[1], index[5] and index[7]: Glucose, BMI and Age.
When to use Information Gain
When the data type of our feature to be tested and the target variable are both categorical.
Summary: Filter Approach
It is possible that different filter-based feature selection methods will return distinct subsets of top n best
features.
Variance threshold: [Insulin]
Correlation coefficient: [Glucose, BMI, Age]
Chi-squared: [Glucose, Insulin, Age]
Information gain: [Glucose, BMI, Age]
To identify determine which feature subset works the best, we can then proceed to examine the performance of
the different subsets on the test set.
Wrapper Approach
We will implement Recursive Feature Elimination (RFE) which is a type of wrapper feature selection method. RFE
works by recursively removing attributes and building a model on those attributes that remain. RFE selects
features by recursively considering smaller and smaller sets of features (backward selection). It uses the model
accuracy to identify which attributes (and combination of attributes) contribute the most to predicting the target
attribute.
To find out more details on the parameters of RFE, check out: https://scikitlearn.org/stable/modules/generated/sklearn.feature_selection.RFE.html#sklearn.feature_selection.RFE
In [9]:
# Import RFE and machine learning algorithm
from sklearn.feature_selection import RFE
# Import SVM - We are using SVM as an example
from sklearn.svm import SVC
# Create a SVM classifier with linear kernel
svmlinear = SVC(kernel = 'linear')
# Use RFE to rank features and return top 3 features
# Parameter step corresponds to the (integer) number of features to remove at each iterati
rfe = RFE(estimator = svmlinear, n_features_to_select = 3, step = 1)
rfe.fit(x_train, y_train)
print("Number of Features: ", rfe.n_features_)
print("Feature Ranking: ", rfe.ranking_)
print("Selected Features: ", rfe.support_)
Number of Features: 3
Feature Ranking: [1 2 4 5 6 1 1 3]
Selected Features: [ True False False False False
True
True False]
The selected features are marked True in the support array and marked with a choice “1” in the ranking array.
The ranking array indicates the strength of these features.
[0] Pregnancies
[1] Glucose
[2] BloodPressure
[3] SkinThickness
[4] Insulin
[5] BMI
[6] DiabetesPedigreeFunction
[7] Age
From the results above, the top 3 features are Pregnancies, BMI and DiabetesPedigreeFunction.
RFE (SVM Linear Kernel): [Pregnancies, BMI, DiabetesPedigreeFunction]
Feature Extraction
Principal Component Analysis (PCA)
Principal Component Analysis (PCA) is a linear dimensionality reduction technique that can be utilized for
extracting information from a high-dimensional space by projecting it into a lower-dimensional sub-space.
In [130…
# Import PCA
from sklearn.decomposition import PCA
# Specify the number of components = 2
pca = PCA(n_components = 2)
# Generate the principal components
pca.fit(x_train)
# Transform the training set into principal components
train_pca = pca.transform(x_train)
# Transform the test set into principal components
test_pca = pca.transform(x_test)
# Convert train set into a data frame to make it easier to view
principalDf = pd.DataFrame(data = train_pca, \
columns = ['principal component 1', 'principal component 2'])
principalDf.head()
principal component 1
principal component 2
0
46.852538
-28.453231
1
-83.841176
19.211268
2
599.569091
9.606891
3
51.211001
20.305400
4
-83.989380
1.647829
Out[130…
PCA transforms the 8 features below into 2 principal components above that can be used as new extracted
features representing the data. This reduces the number of features from 8 to 2 to be fed into the machine
learning model for training.
In [124…
# Print the original training set before PCA
x_train.head()
Pregnancies
Glucose
BloodPressure
SkinThickness
Insulin
BMI
DiabetesPedigreeFunction
Age
603
7
150
78
29
126
35.2
0.692
54
118
4
97
60
23
0
28.2
0.443
22
247
0
165
90
33
680
52.3
0.427
23
157
1
109
56
21
135
25.2
0.833
23
468
8
120
0
0
0
30.0
0.183
38
Out[124…
In [145…
# Print explained variance by principal components
print('Explained variance by component: ', pca.explained_variance_ratio_)
Explained variance by component:
[0.89142243 0.059357
]
We can view the percentage of variance explained by each principal component using explained_varianceratio.
The vector array provided by explained_varianceratio indicates that most of the variance is explained by principal
component 1 (89.1%). Principal component 2 explains 5% of variance. It is therefore to conclude that the first
two principal components explain the majority of the variance.
Factor Analysis
Factor Analysis (FA) can be used to search influential underlying factors or latent variables from a set of observed
variables. It helps reduce the number of variables by explaining the variance among the observed variable and
condense a set of the observed variable into the unobserved variable called factors.
In [49]:
# Import FactorAnalysis
from sklearn.decomposition import FactorAnalysis
# Set the number of factors to 4
factor = FactorAnalysis(n_components = 4, random_state = 101).fit(x_train)
# Convert the factor loadings into a data frame
factor_df = pd.DataFrame(factor.components_, columns = x_train.columns)
factor_df
Pregnancies
Glucose
BloodPressure
SkinThickness
Insulin
BMI
DiabetesPedigreeFunction
Age
0
-0.271827
10.712261
1.634843
6.837644
117.384877
1.536430
0.063206
-0.722935
1
-0.574407
-29.883031
-2.818284
1.459739
1.254262
-1.447046
-0.019330
-3.859221
2
-0.422100
0.515181
-19.009104
-4.219335
0.032111
-2.008041
-0.013237
-2.162622
3
-0.311025
0.135564
-1.519511
13.273705
-0.057911
2.614398
0.044057
-2.200015
Out[49]:
After loading the data and having stored all the predictive features, the FactorAnalysis class is initialized with a
request to look for four factors. The data is then fitted. We can explore the results by observing the
components_ attribute, which returns an array containing measures of the relationship between the newly
created factors, placed in rows, and the original features, placed in columns. The values are known as factor
loadings.
At the intersection of each factor and feature, a positive number indicates that a positive proportion exists
between the two; a negative number, instead, points out that they diverge and one is the contrary to the other.
Pay attention to the high factor loadings regardless of the sign (positive or negative).
Method 1 Feature Selection Using Factor Analysis: One method to use factor analysis for feature selection is to
study the relationships between the attributes to the factors. Performing factor analysis on this dataset actually
did not group the original features into factors. For original features in a factor, we can pick one most relevant
feature from a factor and drop the remaining features in the same factor to reduce the number of features.
Think of factor analysis as an intermediate step to identify factors as a new set of features.
In our example, we can try to group the original features into 4 meaningful factors. For example, Insulin is highly
correlated with Factor 1, Glucose is highly correlated with Factor 2, and so on. The factor loadings for
DiabetesPedigreeFunction is too low, which indicates it is uncorrelated with any of the factors so we can possibly
remove it from further consideration.
Method 2 Feature Extraction Using Factor Analysis: The second method is to transform the original data into
factor scores so our original 8 attributes can be transformed into 4 attributes with each attribute representing a
factor.
In [35]:
# Get factor scores for each row of data
x_transformed = factor.fit_transform(x_train)
# Returns factor scores in an array
x_transformed
array([[ 0.38291592, -0.84029596, -0.33905185,
0.36214101],
[-0.70539703, 0.55392437, 0.29562535, 0.53180043],
[ 5.08854146, 0.3295323 , -0.54832708, -1.68408777],
...,
[-0.70644016, 0.64892983, 0.04124787, 0.35466829],
[-0.71025868, 0.93100429, -0.34030281, -1.30005474],
[-0.69202444, -0.76640417, -0.49131209, -1.33382239]])
Out[35]:
In [51]:
# Convert array into a data frame
df_x_transformed = pd.DataFrame(x_transformed)
# Set the index in df_x_transformed to match the index in x_train so can be mapped back to
df_x_transformed.set_index(x_train.index, inplace = True)
# Rename column names
df_x_transformed.columns = ['Factor 1', 'Factor 2', 'Factor 3', 'Factor 4']
# Factor scores are transformed into features
df_x_transformed
Factor 1
Factor 2
Factor 3
Factor 4
603
0.382916
-0.840296
-0.339052
0.362141
118
-0.705397
0.553924
0.295625
0.531800
247
5.088541
0.329532
-0.548327
-1.684088
157
0.444040
0.567956
0.649008
-0.048220
468
-0.698988
-0.160049
3.525566
-0.034909
...
...
...
...
...
763
0.824161
0.952684
-0.539542
1.174097
192
-0.684019
-1.513588
0.400504
-0.836227
629
-0.706440
0.648930
0.041248
0.354668
559
-0.710259
0.931004
-0.340303
-1.300055
684
-0.692024
-0.766404
-0.491312
-1.333822
Out[51]:
614 rows × 4 columns
Factor analysis transforms the 8 features into 4 features (factors) that can be used as new extracted features
representing the data. This reduces the number of features from 8 to 4 to be fed into the machine learning
model for training.