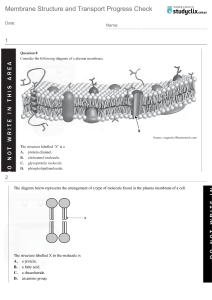
Chapter 1: The Human Body: An Orientation An Overview of Anatomy and Physiology The Language of Anatomy I. Anatomy – study of body organization and I. Directional Terms structure, and their relationships; Structure Superior (cranial) – toward the head determines the function Inferior (caudal) – towards the tail Developmental Anatomy – conception to Anterior (ventral) – towards the belly adulthood Posterior (dorsal) – towards the back Embryology – first 8 weeks of Proximal – near the origin, trunk, or point of development attachment Structures studied with a microscope Distal – farther the origin, trunk, or point of Cytology –cell attachment/ towards the end Histology – tissues Medial – along the coronal towards the Ways to study anatomy sagittal Gross Anatomy – systemic or Lateral – along the coronal away from sagittal regional perspective Superficial – near the surface Surface Anatomy – using Deep – far from the surface anatomical landmarks to locate internal features II. Body Planes Anatomical Imaging – non-invasive Transverse Plane – parallel to the ground way of viewing internal body Coronal Plane – division from front and structures back II. Physiology – the study of the structure’s function Sagittal Plane – division from left and right Exercise physiology – study changes in the III. Body Cavities body after exercise Dorsal – protected by the bone III. Pathology – study of all disorders Cranial – contains the brain Spinal – contains the spinal cord Levels of Structural Organizations Ventral – less protected Thoracic – extends superiorly to the diaphragm; where the heart and lungs are protected by the ribcage Abdominopelvic – extends inferiorly from the diaphragm; contains digestive, urinary, and reproductive organs. The abdomen is only protected by the trunk muscles while the pelvic is somewhat protected by the pelvic bones. Smaller body cavities (Oral, Nasal Orbital, Middle Ear) Maintaining Life Necessary Life Functions: (1) Maintain Boundaries, (2) Movement, (3) Responsiveness, (4) Digestion, (5) Metabolism, (6) Excretion, (7) Reproduction, (8) Growth IV. Regional Terms Homeostasis I. Homeostasis – Ability to regulate internal balances with response to external changes. Disruption can lead to disorder in the body 3 components of homeostatic reflux – (1) receptors sense stimuli > (2) control centers assess and integrate appropriate response to the stimuli > (3) effectors that execute the response (output) II. Negative feedback mechanism – reduce or stop initial stimuli III. Positive feedback mechanism – increase initial stimuli Chapter 2: The Chemical Basis of Life Basic Chemistry III. Chemical Bonding I. Matter, Mass, Weight Matter – is anything that occupies space and has mass. Exists in three forms (solid, liquid, and gas) Mass – amount of matter in a substance or object Weight – exerted force of gravity on matter II. Atoms - Atoms are the basic unit of elements Atoms contain 3 subatomic particles Proton – positive charge (inside the nucleus) Neutron – no charge (inside the nucleus) Electron – negative charge (outside the nucleus) Important values Atomic number - the number of proton/s in an atom. In a neutral atom, the number of electrons is the same as the number of protons Atomic mass unit (amu)/Dalton – the total mass of an atom = number of protons + number of neutrons Isotopes Elements with the same number of protons but different number of neutrons resulting in different atomic mass Electronegativity – degree to which an atom attracts electrons in a chemical bond Ionic bond – characterized as the electrostatic attraction between a positively and negatively charged ion. Between metals and nonmetals (not all) Cation gives up electrons (+) and Anion gains electrons (-) ex. NaCl (Sodium chloride) ED = 2.1 note: metals tend to be cations and nonmetals anion Covalent bond – characterized as the sharing of electrons between non-metals in a chemical bond Polar covalent bond – when atoms don’t have enough electronegativity difference for ionization, the one with higher electronegativity will hog all the electrons (partially negative) from the other (partially positive) ex. HCl (Hydrochloric Acid) ED = 0.9 Valence electrons – the electrons in the outer shell of an atom that will participate in chemical bonding. Octet rule - atoms tend to prefer having 8 electrons in their valence shell. (2-8-8). Non-polar covalent bond – the electrons are shared evenly (typically occurs between atoms of the same element) Moles – standard quantity to represent the 6.02x1023(Avogadro’s number) of small substances (atoms, ions, molecules) IV. Atoms, Molecules, Elements and Compounds ex. salt and sugar dissolved in water VII. Chemical structures Diatomic gases include – Br I N Cl H O F V. Intermolecular Forces – force that mediates interaction between molecules Hydrogen bonding – weak attraction between a positively charged hydrogen and negatively charged oxygen or other polar molecules. ex. (1) water and (2) ethanol Carbons live in ends and bends suffix -ane means single bond suffix -ene means double bond Hydrogen loves carbons ex. caffeine (C8H10N4O2) Fluorine, Oxygen, and Nitrogen can also participate in hydrogen bonding aside from hydrogen Chemical Reactions and Energy VI. Solubility and Dissociation Solubility is a substance’s ability to dissolve in another substance Electrolytes – ionic compounds that dissociate in water and conduct electrical charge Non-electrolytes – molecules that do not dissociate in water and do not create electrical charge I. Synthesis Reaction – A+B=AB (cross multiply the electrical charge then balance) II. Decomposition Reactions III. Single Replacement VII. Energy – ability to do work Potential energy – stored energy that could do work Kinetic Energy – energy doing the work IV. Double Replacement - Metal x Nonmetal V. Reduction-Oxidation Reactions (Redox Reactions) always occur together VI. Reversible Reactions characterized when formed products revert back to their old reactants ex. Ammonium chloride (NH4Cl) reverts back to ammonium (NH3) and Hydrogen chloride (HCl) when heated (thermal decomposition) and both these reactants react to form NH4Cl crystals. Forward – there are more NH4Cl and less NH3; HCl, thus forward is faster Backward – after a while, there will be more NH3; HCl thus, backward rate will be faster Equilibrium – the speed rates of both forward and backward are the same (does not mean one is more) Chemical energy – potential energy stored in the chemical bonds of a substance lesser pt of reactant, energy is required for chemical reaction and the prodcut will have greater pt greater pt of reactant, energy is released for chemical reaction to occur and the product will have greater pt VIII. Speed of Chemical Reactions Activation energy – minimum amount of required energy for chemical reaction to occur Catalyst – substance that speeds up the rate of chemical reaction without it being changed Enzymes – protein catalyst that lowers the activation energy, thus, increasing the speed and rate of chemical reaction note: temperature and concentration can affect the rate of chemical reaction Inorganic Chemistry I. Inorganic Chemistry – are substances that lack carbon or carbon-hydrogen bonds. II. Water – polar molecule that forms hydrogen bonds together or with other polar molecules Polarity/Solvent properties all chemical reactions in the body are dependent on water’s solvent properties acts as a transport and exchange medium as it easily dissolves nutrients, waste products, respiratory gases. Hydrophilic – substances that interact with water (polar molecules and ionic compounds) Hydrophobic – substances that repel water (nonpolar molecules) Cohesion – water molecule – water molecule interaction Adhesion – water molecule – another polar/ionic object Regulates body temp with its high heat capacity. Sweating results from evaporation of water from the body releasing heat Protects body by acting as a lubricant or cushion to avoid friction damage or physical trauma (at joints or body cavities) Acts as a reactant by adding water molecules to chemical bonds to break them down (hydrolysis; water-splitting) III. Electrolytes – produces + charged and – charged ions when dissolved in water (has the ability to conduct electricity in solution) Salts (Na+Cl-) – ionic compound that dissociates in water PH, Acids and Bases Acids – substance that release hydrogen ions (H+) or proton (H+) donor o strong acids – completely ionize and liberate all their protons (ex. HCl H+ + Cl-) o weak acids – ionize incompletely (ex. ) Bases – grabs protons or donates OH- when they dissociate (ex. NaOH Na+ + OH-) or (OH- + H+ H2O) pH – express acidity or alkalinity of solution (enzymes only work at certain pH and a change can result in denaturing) a change in pH by 1 in solution represents a 10-fold change of H+ concentration. The lower the value means H+ is more concentrated, thus, acidic. Buffers – molecule that maintains pH at a constant rate note: Normal blood pH ranges from 7.35 to 7.45. Slight deviations outside this range can be fatal Organic Chemistry I. Organic Chemistry – Carbon containing compounds that comprise living matter. Many of these compounds are polymers (carbohydrate and protein) Polymers - chainlike structures that is formed by a single unit called monomers that are joined together by dehydration synthesis Dehydration synthesis – one hydrogen (H) atom is removed from one monomer and a hydroxyl group (OH) is removed from the other monomer (releasing H2O) Hydrolysis – adding H2O between bonds to release the monomers II. Carbohydrate – it contains carbon-hydrogenoxygen in a 1:2:1 ratio acts as a short term (converted to ATP) and long term (glycogen for animals, starch and cellulose for plants) energy storage suffix -ose is used for describing and naming carbohydrates Monosaccharide – one sugar; the building block of carbohydrate (fruits, veggies, honey) Glucose – blood sugar; universal cellular fuel Fructose and Galactose – converted to glucose for cell function Deoxyribose and Ribose – part of nucleic acid structure Polysaccharides – chains of simple sugars Starch – grains or root vegetables Glycogen – long term energy storage molecule in muscles/liver Cellulose – can’t be used as energy but many nutritional benefits (dietary fibers; leafy greens) III. Lipids – energy rich non-polar (hydrophobic) compounds containing carbon-hydrogen-oxygen in a 1:2: little oxygen ratio (C57H110O6) Triglycerides – one glycerol backbone with three fatty acids linked to it by dehydration synthesis Saturated fats – single C-C bonds pack closely at room temperature Unsaturated fats – double or triple bonds between C-C causing it to curve, thus, not solidify at room temperature Omega-3 and Omega-6 polyunsaturated - heart healthy the first double bond is found at the 3rd and 6th carbon Disaccharide – two monosaccharides joined by dehydration synthesis and is broken down by hydrolysis for digestion Sucrose: Table sugar Lactose: Milk Maltose: Malt sugar (beer) Trans Fats - chemically altered unsaturated fat by hydrogenation (unsuccessful) (rare in nature, thus, no enzyme to break it down) The structure and function of protein is dictated by the type and order of amino acids Phospholipids – same to triglycerides except that one of the fatty acids is replaced by a phosphate containing region. the polar side is the one containing the phosphate and the nonpolar side which contains the fatty acids Steroids – do not contain fatty acids, instead, have interlocking four carbon ring structure. Cholesterol – found in cell membranes and synthesize other steroids Bile salts – increase fat absorption Reproductive hormones – testosterone and estrogen for secondary sex characteristics Peptide – 2 amino acids bonded by dehydration or protein synthesis Dipeptide 2 amino acids joined together Tripeptide 3 amino acids joined together Tetrapeptide 4 amino acids joined together Oligopeptide 2-20 amino acids Polypeptide 21-49 amino acids Protein More than 50 amino acids Four Levels of Protein Structure Primary – sequence of amino acids in the polypeptide chain determined by DNA IV. Proteins Amino Acids – monomers of proteins Secondary - hydrogen bonding (dotted red lines). The hydrogen bonds cause the amino acid chain to form pleated (folded) sheets or helices (coils). Tertiary – interaction of side chains (denaturing occurs when these interactions break down) Quaternary – formed between the interaction of 2 or more polypeptide units and can be globular or fibrous V. Nucleic Acids – organic living substance found in cells that can either be a DNA or RNA polymer made up of nucleotides as monomers (basic unit) • Nucleotides – has 3 basic parts Pentose sugar – deoxyribose (no O on carbon 2) for DNA and ribose (OH in carbon 2) for RNA Nitrogenous bases > Purine (2 rings) – sugar ring will attach to nitrogenous base at 9th position > Pyrimidine (1 ring) - sugar ring will attach to nitrogenous base at 1st position Nucleoside (sugar and base only) Phosphate group (PO-34) • DNA (deoxyribonucleic acid) - determines the structure and sequence of amino acids - store genetic information Functional unit is nucleotide [phosphate group bonds (covalent) to deoxyribose at carbon 5, deoxyribose is also bonded to 1 nitrogenous base] P-group of another nucleotide connects to carbon 3, making the sugar-phosphate backbone of DNA and sequence of the bases which are now codes [P-group is now bonded to two sugars (3’5’ phosphodiester bonds)] Complementary base pairs are present on the other strand which creates a hydrogen bond between the bases (2 bonds between AT and 3 bonds between GC) The complementary strands are antiparallel in that the 5' > 3' direction of one strand runs counter to the 5' > 3' direction of the other strand. The nucleotide strands coil to form a double-stranded helix note: some DNA are non-coding, some makes genes that aren’t activated, and some makes genes that code for active protein • RNA (ribonucleic acid) single stranded functional unit is nucleotide (Pgroup attached to carbon 5 of ribose, that is also attached to a nitrogenous base) and also forms 3’5’ phosphodiester bonds nitrogenous base – AU and GC 3 types – messenger RNA, ribosomal RNA, and transfer RNA • Protein Synthesis (1st level of protein structure) Transcription – RNA polymerase binds complementary RNA bases to DNA template strand in nucleus to create mRNA based on DNA sequence mRNA goes out of nucleus and attach to ribosome; rRNA is a component of ribosome Translation – tRNA brings amino acids based on the anticodon tRNA has and finds the complementary bases on mRNA by reading it by 3 (codon) tRNA leaves the ribosome and leave the amino acid behind which bond with other amino acids. Naming nucleosides/tides AMP (adenine+ribose+monophosphate) ADP (adenine+ribose+diphosphate) ATP (adenine+ribose+triphosphate) - close proximity of 3 (-) charged P-groups are unstable, thus, stores high amount of PE - when phosphate is released (hydrolysis) small amount of energy is released, ATP > ADP - chemical reactions like catabolism of glucose or other nutrient release energy to synthesize ADP > ATP - when ATP is produced, it is used for functions that require energy like anabolism or propel cell function (muscle contraction); ATP > ADP Chapter 3: Cell Biology Introduction to Cells I. Cell Theory 1. All living organisms are made up of cells 2. Cell is the basic structural and functional unit of life 3. All cells arise from pre-existing cells Important Cell Milestones II. Parts and Functions of the Cell Note: Cell size and shape vary depending on their functions • Plasma Membrane/Cell Membrane Transparent fluid barrier that separates the cell from its environment Semi-permeable/selectively permeable - allows only specific substances or molecules to enter the cell Lipid-soluble/non-polar molecules can easily pass while polar molecules cannot ECF (extracellular fluid) - high levels of sodium - low levels of potassium ICF (intracellular fluid) - high levels of potassium - low levels of sodium Fluid-Mosaic Model - represents the biological structure of a plasma membrane along its components that determine its fluid-like nature Plasma Membrane/Cell Membrane > Transparent fluid barrier that separates the cell from its environment > Semi-permeable/selectively permeable - allows only specific substances or molecules to enter the cell > Lipid-soluble/non-polar molecules can easily pass while polar molecules cannot > ECF (extracellular fluid) - high levels of sodium - low level of potassium + calcium a + chloride ions > ICF (intracellular fluid) - high levels of potassium - low levels of sodium +enzymes +proteins +glycogen + potassium ions Fluid-Mosaic Model - represents the biological structure of a plasma membrane along its components that determine its fluid-like nature Membrane Lipids > Polar heads oriented to face the ICF and ECF > Nonpolar Phospholipid > Hydrophilic, polar head (phosphate group + R (choline) > Hydrophobic, non-polar tail composed of two fatty acids (saturated and unsaturated) Cholesterol > Maintains fluidity of the phospholipid bilayer: 1) preventing it to solidify in low temperatures by pushing them apart and 2) preventing it to be too fluid in high temperatures by holding the phospholipids together. Fluid-mosaic model > Explains that the plasma membrane is fluid-like (like a dense liquid) and is neither static nor rigid > Flexible, thus, can change shape and composition through time Membrane Proteins and their Functions > Integral proteins - proteins that are embedded within the phospholipid bilayer > Peripheral proteins - proteins that attach to either the inner or outer surfaces of the bilayer > Surface proteins - proteins that lies on inner or outer cell surface Marker Molecules Helps the cell recognize other cells (Carbohydrate) Glycoprotein - carbohydrate attached to protein Glycolipids - carbohydrates attached to lipid Ex. 1. sperm cell recognizing oocyte 2. white blood cells recognizing foreign invaders (distinguishing bacteria from donor cells) Attachment Proteins ○ Cadherins > Proteins that attach to the protein of another cell ○ Integrins > Proteins that attach with other molecules from inside and outside the cell Transport Proteins > Integral proteins that extend from each side of the membrane (act as a passageway of substances) Three common characteristics: a) Specificity - substances can only enter through a protein specific to the substance b) Competition - substances with the same shape bind to a common protein (substances in higher concentration are transported at a faster rate) c Saturation - substances that get transported are limited by the number of transport proteins. ○ Channel Proteins > solutes don't bind to channel proteins ■ Leak Ion Channels A Passive Transport Protein > always open for transport of specific substances ■ Gated Ion Channels ● Ligand Gated Ion Channel > Ligand (chemical signal) binds with the protein receptor to open ● ○ Voltage-Gated Ion Channel > Gates open in response to change in membrane potential Carrier Proteins > Proteins bind to protein receptors which triggers a conformational change ■ Uniport > Carries one specific substance at a time ■ Symport Cotransport > Transports two different substances together in the same direction ■ Antiport Counter-transport > transports one substance to a different side, then transport a different substance back to the first side ○ ATP-Powered Pumps > Have 2 binding sites > One for the ion to be transported and one for ATP which will release energy to fuel the transport Receptor Proteins > Proteins with exposed specific receptors wherein chemical signals (or other substances) bind to initiate a response > can be membrane proteins or glycoproteins ○ Receptor Linked to Channel Proteins > channel proteins have an exposed receptor wherein chemical signals bind to change ○ Receptors Linked to G Protein Complexes > the receptor site of a protein binds with a chemical signal which causes the G protein complex to bind to the protein. > the guanosine diphosphate from the a-subunit of the complex will then be replaced with guanosine triphosphate (activation) > the g-complex leaves the protein and the a-subunit will then dissociate from the complex to be used for cell Enzymes > are integral proteins that catalyze chemical reactions from the inner or outer surface of the cell > are either always active or activated by G protein complexes Movement Through the Plasma Membrane Passive Membrane Transport > does not require energy (substance moves along the concentration gradient which allows substances to move from high to low concentration) > collisions from concentrated area creates energy ○ Diffusion > movement (high-to-low concentration) of lipid-soluble substances directly through the phospholipid bilayer to the opposite side of the membrane > small polar molecules can diffuse (ex. water) > water diffusion is called osmosis ○ Osmosis > movement of water from high to low concentration (requires aquaporins) ■ Tonicity > behavior of cells when placed in a solution (shrink or expand and burst) > 3 types of solutions ● Hypotonic > hypo[less]+tonic[solute] > less solute, thus more water > cell has more solute and less water > water moves from solution to cell (expand and burst) ● Hypertonic > hyper[high]+tonic[solute] > more solute, thus, less water > cell has less solute and more water > water moves from cell to solution (shrink) ● ○ Isotonic > concentration of solute and solvent is the same throughout the sides, thus, no net movement (no gradient) Facilitated Diffusion > movement of large lipid-insoluble (hydrophilic, polar) substances across the membrane through the help of transmembrane proteins > specificity, competition, and saturation Active Membrane Transport > requires energy (ATP) for substances to move against the gradient or from low to high concentration ○ Active Transport > transport of inorganic or ionic substances through ATP-powered pumps (require energy) from high to low concentration or vice versa > ex.: In the ECF, Na is higher and K is lower which means in ICF, Na is lower and K is higher. In a sodium-potassium pump, K moves to ICF (low to high) and Na moves to ECF (low to high) > from ICF, sodium attaches to ATP pump, when ATP binds to pump and breaks down, it undergoes a conformational change which pumps sodium to ECF > K attaches to the pump from ECF, when phosphate is released, it goes back to its original shape, thus, releasing the K to ICF note: for every ATP, Na moves to ECF and K moves from ECF back to ICF ○ Secondary Active Transport > movement of organic or ionic substances (glucose and Na) along and against the concentration gradient > ex.: when Na accumulates in the ECF, it moves down to its gradient which creates energy for Glucose to move inside the cell (against the gradient) Vesicular Transport > transport of large substances across the membrane > requires ATP ○ Endocytosis > moves substances from outside to inside the cell ■ Phagocytosis > phago (to eat) > cell-eating > ex.: macrophage (a wbc) finds a streptococcus (a bacterium) > strep attaches to macro receptors > macro extends pseudopods across the strep to envelope > vesicle is formed then separated from the membrane (phagosome) > phagosome meets lysosome > lysosome digests phago with its digestive enzymes > leftovers are expelled note: phagocyte is an immune cell ■ Pinocytosis > cell drinking > membrane engulfs random molecules and fluid from ICF and delivers it deep into the cytosol where it is released ■ Receptor-Mediated Endocytosis > are special receptors that carry specific into the cell to increase cell uptake on these specific substances > ex. LDL (low-density lipoprotein) receptor is found in clathrin-coated pits in the cell membrane > LDL bind to the receptors > pseudopods form then clathrins link up around it and gets released back to the membrane when vesicle separates from the membrane > endosome engulfs the vesicle and separated LDL from the receptor > 2 vesicles form for LDL and receptor > LDL goes to the lysosome for digestion > receptor goes back to membrane > when LDL receptor is low, cholesterol uptake of the cell is low thus, cells produce more cholesterol inside (hypercholesterolemia) when cholesterol accumulates it blocks blood vessels (Atherosclerosis) which causes heart attack or stroke ○ Exocytosis > ICF substances expelled out of the cell ○ > ex.: Golgi apparatus packs proteins, lipids, and hormones from the endoplasmic reticulum through a vesicle > vesicle is transported through cytoskeleton to the membrane > vesicular membrane fuse with cell membrane > vesicular contents rupture to the ECF Cell Junctions > cells need to connect and communicate with each other through different types of junctions Tight Junctions > holds cell very tightly where waters and ions can't pass through with the help of proteins > ex. bladder, intestine, stomach, kidney, etc. Desmosomes > connects cell through cadherins (proteins) that extends to the cytoskeleton > waters and ions can pass through for flexibility and reduced pressure > skin, heart muscle, and intestine Gap Junctions > creates a connection between cells (connexon) for intercellular communication (ions and molecules pass through or spread action potential) Plasma Membrane/Cell Membrane > Transparent fluid barrier that separates the cell from its environment > Semi-permeable/selectively permeable - allows only specific substances or molecules to enter the cell > Lipid-soluble/non-polar molecules can easily pass while polar molecules cannot > ECF (extracellular fluid) - high levels of sodium - low level of potassium + calcium a + chloride ions > ICF (intracellular fluid) - high levels of potassium - low levels of sodium +enzymes +proteins +glycogen + potassium ions Fluid-Mosaic Model represents the biological structure of a plasma membrane along its components that determine its fluidlike nature Membrane Lipids > Polar heads oriented to face the ICF and ECF > Nonpolar Phospholipid > Hydrophilic, polar head (phosphate group + R (choline) > Hydrophobic, non-polar tail composed of two fatty acids (saturated and unsaturated) Cholesterol > Maintains fluidity of the phospholipid bilayer: 1) preventing it to solidify in low temperatures by pushing them apart and 2) preventing it to be too fluid in high temperatures by holding the phospholipids together. Fluid-mosaic model > Explains that the plasma membrane is fluid-like (like a dense liquid) and is neither static nor rigid > Flexible, thus, can change shape and composition through time Membrane Proteins and their Functions > Integral proteins - proteins that are embedded within the phospholipid bilayer > Peripheral proteins proteins that attach to either the inner or outer surfaces of the bilayer > Surface proteins - proteins that lies on inner or outer cell surface Marker Molecules Helps the cell recognize other cells (Carbohydrate) Glycoprotein - carbohydrate attached to protein Glycolipids - carbohydrates attached to lipid Ex. 1. sperm cell recognizing oocyte 2. white blood cells recognizing foreign invaders (distinguishing bacteria from donor cells) Attachment Proteins ○ Cadherins > Proteins that attach to the protein of another cell ○ Integrins > Proteins that attach with other molecules from inside and outside the cell Transport Proteins > Integral proteins that extend from each side of the membrane (act as a passageway of substances) Three common characteristics: a) Specificity - substances can only enter through a protein specific to the substance b) Competition - substances with the same shape bind to a common protein (substances in higher concentration are transported at a faster rate) c) Saturation substances that get transported are limited by the number of transport proteins. ○ ○ Channel Proteins > solutes don't bind to channel proteins ■ Leak Ion Channels A Passive Transport Protein > always open for transport of specific substances ■ Gated Ion Channels ● Ligand Gated Ion Channel > Ligand (chemical signal) binds with the protein receptor to open ● Voltage-Gated Ion Channel > Gates open in response to change in membrane potential Carrier Proteins > Proteins bind to protein receptor which triggers conformational change ■ Uniport > Carries one specific substance at a time ○ ■ Symport Cotransport > Transports two different substances together in the same direction ■ Antiport Counter-transport > transports one substance to a different side, then transport a different substance back to the first side ATP-Powered Pumps > Have 2 binding sites > One for the ion to be transported and one for ATP which will release energy to fuel the transport Receptor Proteins > Proteins with exposed specific receptors wherein chemical signals (or other substances) bind to initiate a response > can be membrane proteins or glycoproteins ○ Receptor Linked to Channel Proteins > channel proteins have an exposed receptor wherein chemical signals bind to change ○ Receptors Linked to G Protein Complexes > the receptor site of a protein binds with a chemical signal which causes the G protein complex to bind to the protein. > the guanosine diphosphate from the a subunit of the complex will then be replaced with guanosine triphosphate (activation) > the g-complex leaves the protein and the a subunit will then dissociate from the complex to be used for cell Enzymes > are integral proteins that catalyze chemical reactions from the inner or outer surface of the cell > are either always active or activated by G protein complexes Movement Through the Plasma Membrane Passive Membrane Transport > does not require energy (substance moves along the concentration gradient which allows substances to move from high to low concentration) > collisions from concentrated area creates energy ○ Diffusion > movement (high-to-low concentration) of lipid-soluble substances directly through the phospholipid bilayer to the opposite side of the membrane > small polar molecules can diffuse (ex. water) > water diffusion is called osmosis ○ Osmosis > movement of water from high to low concentration (requires aquaporins) ■ Tonicity > behavior of cells when placed in a solution (shrink or expand and burst) > 3 types of solutions ● Hypotonic > hypo[less]+tonic[solute] > less solute, thus more water > cell has more solute and less water > water moves from solution to cell (expand and burst) ○ ● Hypertonic > hyper[high]+tonic[solute] > more solute, thus, less water > cell has less solute and more water > water moves from cell to solution (shrink) ● Isotonic > concentration of solute and solvent is the same throughout the sides, thus, no net movement (no gradient) Facilitated Diffusion > movement of large lipid-insoluble (hydrophilic, polar) substances across the membrane through the help of transmembrane proteins > specificity, competition, and saturation Active Membrane Transport > requires energy (ATP) for substances to move against the gradient or from low to high concentration ○ Active Transport > transport of inorganic or ionic substances through ATP-powered pumps (require energy) from high to low concentration or vice versa > ex.: In the ECF, Na is higher and K is lower which means in ICF, Na is lower and K is higher. In a sodium-potassium pump, K moves to ICF (low to high) and Na moves to ECF (low to high) > from ICF, sodium attaches to ATP pump, when ATP binds to pump and breaks down, it undergoes a conformational change which pumps sodium to ECF > K attaches to the pump from ECF, when phosphate is released, it goes back to its original shape, thus, releasing the K to ICF note: for every ATP, Na moves to ECF and K moves from ECF back to ICF ○ Secondary Active Transport > movement of organic or ionic substances (glucose and Na) along and against the concentration gradient > ex.: when Na accumulates in the ECF, it moves down to its gradient which creates energy for Glucose to move inside the cell (against the gradient) Vesicular Transport > transport of large substances across the membrane > requires ATP ○ Endocytosis > moves substances from outside to inside the cell ■ Phagocytosis > phago (to eat) > cell-eating > ex.: macrophage (a wbc) finds a streptococcus (a bacterium) > strep attaches to macro receptors > macro extends pseudopods across the strep to envelope > vesicle is formed then separated from the membrane (phagosome) > phagosome meets lysosome > lysosome digests phago with its digestive enzymes > leftovers are expelled note: phagocyte is an immune cell ■ Pinocytosis > cell drinking > membrane engulfs random molecules and fluid from ICF and delivers it deep into the cytosol where it is released ■ ○ Receptor-Mediated Endocytosis > are special receptors that carry specific into the cell to increse cell uptake on these specific substances > ex. LDL (low-density lipoprotein) receptor is found in clathrin-coated pits in the cell membrane > LDL bind to the receptors > pseudopods form then clathrins link up around it and gets released back to the membrane when vesicle separates from the membrane > endosome engulfs the vesicle and separated LDL from the receptor > 2 vesicles form for LDL and receptor > LDL goes to lysosome for digestion > receptor goes back to membrane > when LDL receptor is low, cholesterol uptake of cell is low thus, cells produce more cholesterol inside (hypercholesterolemia) when cholesterol accumulates it blocks blood vessels (Atherosclerosis) which causes heart attack or stroke Exocytosis > ICF substances expelled out of the cell > ex.: Golgi apparatus packs proteins, lipids, and hormones from the endoplasmic reticulum through a vesicle > vesicle is transported through cytoskeleton to the membrane > vesicular membrane fuse with cell membrane > vesicular contents rupture to the ECF Cell Junctions > cells need to connect and communicate with each other through different types of junctions Tight Junctions > holds cell very tightly where waters and ions can't pass through with the help of proteins > ex. bladder, intestine, stomach, kidney, etc. Desmosomes > connects cell through cadherins (proteins) that extends to the cytoskeleton > waters and ions can pass through for flexibility and reduced pressure > skin, heart muscle, and intestine Gap Junctions > creates a connection between cells (connexon) for intercellular communication (ions and molecules pass through or spread action potential)