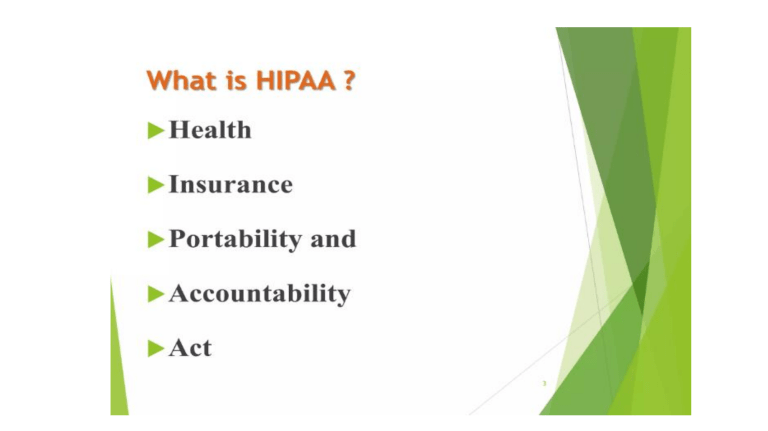
Translational research informatics Translational Research as defined by the National Institutes of Health includes two areas of translation. One is the process of applying discoveries generated during research in the laboratory, and in preclinical studies, to the development of trials and studies in humans. The second area of translation concerns research aimed at enhancing the adoption of best practices in the community. Cost-effectiveness of prevention and treatment strategies is also an important part of translational research. Unit 5 Legislation and Regulation: Health Insurance Portability and Accountability Act, Certification Commission for Healthcare Information Technology, Software Systems, Medical software, Dental software, List of freeware health software, List of open source healthcare software, List of neuroimaging software, Mirth, Mpro , Open Dental, Personal Health Application . Clinical Research Informatics: Translational research informatics, Clinical trial management, Clinical data management system, Case report form, Clinical coder, Clinical data acquisition, Data clarification form, Patientreported outcome. Standards, Coding and Nomenclature: Diagnosis codes, Procedure codes. Legislation and Regulation Health Insurance Portability and Accountability Act (HIPAA) Bill Clinton in 1996 Title I of HIPAA protects health insurance coverage for workers and their families when they change or lose their jobs. Title II of HIPAA, known as the Administrative Simplification (AS) provisions, requires the establishment of national standards for electronic health care transactions and national identifiers for providers, health insurance plans, and employers The administrative simplification provisions also address the security and privacy of health data. The standards are meant to improve the efficiency and effectiveness of the nation's health care system by encouraging the widespread use of electronic data interchange in the U.S. health care system Image processing software Free • 3DSlicer • AFNICell • Cognition • CellProfiler • DlibEndrov • FijiF • MRIB • Software Library • Free Surfer • GemIdentGNU Octaveilastik • ImageJ • ITKInVesaliusITKSNAPKNIMEMangoOpenCVOsiriXVIGRAVXL Proprietary • • • • • • • • • • Amira Analyze Aphelion Bitplane IDL Mathematica MATLAB Mimics MountainsMap Visage SDK Analyze (imaging software) Analyze is a software package developed by the Biomedical Imaging Resource (BIR) at Mayo Clinic for multi-dimensional display, processing, and measurement of multi-modality biomedical images. It is a commercial program and is used for medical tomographic scans from magnetic resonance imaging, computed tomography and positron emission tomography. The Analyze 7.5 file format[1] has been widely used in the functional neuroimaging field, and other programs such as SPM, FreeSurfer, AIR, MRIcro and Mango are able to read and write the format. The files can be used to store voxel-based volumes. One data item consists of two files: One file with the actual data in a binary format with the filename extension .img and another file (header with filename extension .hdr) with information about the data such as voxel size and number of voxels in each dimension. SPM has defined changes to this format, among other things the voxel ordering within the file. SEGMENTATION •Interactive volume segmentation •Semi-automatic object segmentation •Comprehensive manual segmentation •Dual-input segmentation •Surface generation TRANSFORM •Spatial transformations such as cropping and flipping •Interactive orthogonal and oblique reslicing •Intensity-based transformations •Mathematical processing •Image correction 3D Slicer 3DSlicer 3D Slicer (Slicer) is a free, open source software package for image analysis and scientific visualization. Slicer is used in a variety of medical applications, including autism, multiple sclerosis, systemic lupus erythematosus, prostate cancer, schizophrenia, orthopedic biomechanics, COPD, cardiovascular disease and neurosurgery. Slicer's capabilities include: • Handling DICOM images and reading/writing a variety of other formats • Interactive visualization of volumetric Voxel images, polygonal meshes, and volume renderings • Manual editing • Fusion and co-registering of data using rigid and non-rigid algorithms • Automatic image segmentation • Analysis and visualization of diffusion tensor imaging data • Tracking of devices for image-guided procedures. What is FreeSurfer? • A suite of software tools for the analysis of neuroimaging data • Full characterizes anatomy – Cortex – thickness, folding patterns, ROIs – Subcortical – structure boundaries • Surface-based inter-subject registration • Multi-modal integration – fMRI (task, rest, retinotopy) – DTI tractography – PET, MEG, EEG Why is FreeSurfer special? • There are other cortical and subcortical tools: – BrainVoyager, Caret, BrainVisa, SPM, FSL (of late) • Each has varying degrees of segmentation accuracy w/ varying levels of user intervention • FreeSurfer is highly specialized in it’s: – cortical surface representation from the grey matter segmentation – surface-based group registration capabilities – accuracy of subcortical structure measurements Why FreeSurfer? • Anatomical analysis is not like functional analysis – it is completely stereotyped. • Registration to a template (e.g. MNI/Talairach) doesn’t account for individual anatomy. • Even if you don’t care about the anatomy, anatomical models allow functional analysis not otherwise possible. FreeSurfer ImageJ • An adaptation of NIH image for the Java platform. • Can run on any computer systems that can run Java (Sun Microsystems) • Open source • Two powerful scripting languages • Java Plugins • Macro Language • Continual Upgrades • Active community of several thousand users The Image Histogram Log Scale The histogram shows the number of pixels of each value, regardless of location. The log display allows for the visualization of minor components. Note that there are unused pixel values Brightness Adjustment The brightness adjustment essentially adds or subtracts a constant to every pixel, causing a shift in the histogram along the x axis, but no change in the distribution Contrast Enhancement For contrast enhancement, a lower value, in this case, 88, is set at zero, and a higher value, 166, is set at 255. The values of each of the pixels are adjusted proportionately. Note that because of the integer values, not all of the pixel values are used. Medical and biological signal applications Medical monitor In medicine, monitoring is the observation of a disease, condition or one or several medical parameters over time Cardiac monitoring • Continuous electrocardiography • Holter monitor • Invasive Swan-Ganz catheter Hemodynamic monitoring • blood pressure and blood flow • invasively through an inserted blood pressure transducer • noninvasively with an inflatable blood pressure cuff Respiratory monitoring • Pulse oximetry which involves measurement of the saturated percentage of oxygen in the blood, referred to as SpO2, and measured by an infrared finger cuff • Capnography, which invoolves CO2 measurements, referred to as EtCO2 or end-tidal carbon dioxide concentration. • The respiratory rate monitored as such is called AWRR or airway respiratory rate) Neurological monitoring • intracranial pressure • electroencephalography, gas anesthetic concentrations, bispectral index (BIS), etc. • brain EEG monitors have a larger multichannel capability Blood glucose monitoring Childbirth monitoring Body temperature monitoring through an adhesive pad containing a thermoelectric transducer Components Sensor - biosensors and mechanical sensors Translating component Display device Physiological data are displayed continuously on a CRT, LED or LCD screen numerical readouts such as maximum, minimum and average values, pulse and respiratory frequencies, and so on Communication links An anesthetic machine with integrated systems for monitoring of several vital parameters, including blood pressure and heart rate. Mobile appliances Applications Blood glucose monitoring Stress monitoring Bio sensors may provide warnings when stress levels signs are rising before human can notice it and provide alerts and suggestions. Serotonin biosensor Future serotonin biosensors may assist with mood disorders and depression. Continuous blood test based nutrition In the field of evidence-based nutrition, a lab-on-a-chip implant that can run 24/7 blood tests may provide a continuous results and a computer can provide nutrition suggestions or alerts. Psychiatrist-on-a-chip In clinical brain sciences drug delivery and in vivo Bio-MEMS based biosensors may assist with preventing and early treatment of mental disorders Epilepsy monitoring In epilepsy, next generations of long-term video-EEG monitoring may predict epileptic seizure and prevent them with changes of daily life activity like sleep, stress, nutrition and mood management. Toxicity monitoring Smart biosensors may detect toxic materials such mercury and lead and provide alerts Wireless Capsule Endoscopy The PASCAL Dynamic Contour Tonometer Holter monitor Holter is a portable device for continuously monitoring various electrical activity of the cardiovascular system for at least 24 hours (often for two weeks at a time). The Holter monitor is named after physicist Norman J. Holter Atrial fibrillation recorded by a Holter monitor Canine Holter Monitor with DogLeggs Vest A Holter monitor can be worn for many days without causing significant discomfort. Automated ECG interpretation Automated ECG interpretation is the use of artificial intelligence and pattern recognition software and knowledge bases to carry out automatically the interpretation, test reporting and computer-aided diagnosis of electrocardiogram tracings obtained usually from a patient. ECG Waveform Phases 1. A digital representation of each recorded ECG channel is obtained, by means of an analogdigital conversion device and a special data acquisition software or a digital signal processing (DSP) chip. 2. The resulting digital signal is processed by a series of specialized algorithms, which start by conditioning it, e.g., removal of noise, baselevel variation, etc. 3. Feature extraction: mathematical analysis is now performed on the clean signal of all channels, to identify and measure a number of features which are important for interpretation and diagnosis, this will constitute the input to AI-based programs, such as the peak amplitude, area under the curve, displacement in relation to baseline, etc., of the P, Q, R, S and T waves, the time delay between these peaks and valleys, heart rate frequency (instantaneous and average), and many others. Some sort of secondary processing such as Fourier analysis and wavelet analysis may also be performed in order to provide input to pattern recognition-based programs. 4. Logical processing and pattern recognition, using rule-based expert systems, probabilistic Bayesian analysis or fuzzy logics algorithms, cluster analysis, artificial neural networks, genetic algorithms and others techniques are used to derive conclusions, interpretation and diagnosis 5. A reporting program is activated and produces a proper display of original and calculated data, as well as the results of automated interpretation. 6. In some applications, such as automatic defibrillators, an action of some sort may be triggered by results of the analysis, such as the occurrence of an atrial fibrillation or a cardiac arrest, the sounding of alarms in a medical monitor in intensive-care unit applications, and so on. MECIF Protocol The MECIF Protocol (Medical Computer Interface Protcol), is a rare communications protocol originally developed by Hewlett-Packard to allow external devices (e.g. computers) to communicate with certain Hewlett-Packard patient monitors. It is a client–server based protocol that uses a modified RS-232 cable to allow a client (e.g. a computer) to send commands to a server (e.g. patient monitor). The protocol can be used to retrieve vital data from patient monitors, such as ECG, blood pressure and heart-rate signals. SCP-ECG SCP-ECG, which stands for Standard communications protocol for computer assisted electrocardiography, is a standard for ECG traces, annotations, and metadata, that specifies the interchange format and a messaging procedure for ECG cart-to-host communication and for retrieval of SCP-ECG records from the host to the ECG cart. It is defined in the joint ANSI/AAMI standard EC71:2001 and in the CEN standard EN 1064:2005. History The SCP Standard was first developed between 1989 to 1991 during a European AIM R&D project. External links "OpenECG" — The [OpenECG] Group supports SCP-ECG by providing and supporting open source implementations and consistent application the standard. Other ECG data formats DICOM, HL7 aECG European Data Format European Data Format (EDF) is a standard file format designed for exchange and storage of medical time series. Being an open and non-proprietary format, EDF(+) is commonly used to archive, exchange and analyse data from commercial devices in a format that is independent of the acquisition system. EDF was published in 1992 and stores multichannel data, allowing different sample rates for each signal. Internally it includes a header and one or more data records. The header contains some general information (patient identification, start time...) and technical specs of each signal (calibration, sampling rate, filtering, ...), coded as ASCII characters. The data records contain samples as little-endian 16-bit integers. EDF+ was published in 2003. EDF+ has applications in PSG, electroencephalography (EEG), electrocardiography (ECG), electromyography (EMG), and Sleep scoring. EDF+ can also be used for nerve conduction studies, evoked potentials and other data acquisition studies OpenXDF The Open eXchange Data Format, or OpenXDF, is an open, XML-based standard for the digital storage and exchange of time-series physiological signals and metadata. OpenXDF primarily focuses on electroencephalography and polysomnography. History Neurotronics began work on OpenXDF in 2003 with the goal of providing a modern, open, and extensible file format with which clinicians and researchers can share physiological data and metadata, such as signal data, signal montages, patient demographics, and event logs.[citation needed] Neurotronics released the first draft of the OpenXDF Specification just before the 18th meeting of the Associated Professional Sleep Societies in 2004. Neurotronics has since relinquished control of the format to the OpenXDF Consortium. As of version 1.0, OpenXDF is 100% backward compatible with the European Data Format (EDF), the current defacto standard format for physiological data exchange