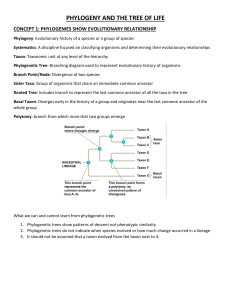
Chapter 16 Notes http://notetaker.top/ 16.1 All of Life Is Connected through Its Evolutionary History The sequencing of complete genomes from various species has confirmed that all of life is connected through a common ancestor. Phylogenetic trees are used to depict evolutionary history of species, populations, and genes. Phylogenetic trees have been constructed from physical structures, behaviors, and biochemical attributes. Phylogenetic trees are used to represent historical lineage splits and the evolution of new traits. Phylogenetic Trees A phylogenetic tree is a diagrammatic reconstruction of the genealogical relationships between species. The tree of life is used to organize biological classification. The positions of the nodes on the time scale indicate the times of divergence events. Taxon (plural taxa) are any group of species designated with a name. Clades are any taxon that consists of all the evolutionary descendants of a common ancestor. Sister species/clades are any two species/clades that are each others' closest relatives. Phylogenetic trees are widely used in molecular biology, biomedicine, physiology, behavior, ecology, and virtually all other fields of biology. Traits and Evolutionary Reversals Any features shared by two or more species that have been inherited from a common ancestor are homologous traits. Similar traits in unrelated groups of organisms can develop by convergent evolution. A character may also revert from a derived state back to an ancestral state in an event called an evolutionary reversal, resulting in homoplastic traits. Derived traits that are shared among a group of organisms and are also viewed as evidence of the common ancestry of the group are synapomorphies. 16.2 Phylogeny Can Be Reconstructed from Traits of Organisms Assumptions Made Traits only arose once during evolution No derived traits were lost from any descendant groups Ingroup vs Outgroup Ingroup: organisms being compared in a phylogenetic study Outgroup: closely related species or group known to be outside the group of interest Lamprey is used as the outgroup for the eight vertebrate animals studied Synapomorphies and Homoplasies Synapomorphies: derived traits that evolved within the ingroup Homoplasies: traits that evolved separately in different groups Parsimony principle: preferred explanation of observed data is the simplest explanation Occam's razor: simplest explanation is the best explanation Phylogenetic trees are continually modified as additional information is obtained Mathematical Models DNA sequences can be used to infer phylogenies Mathematical models account for multiple changes at a given position in a DNA sequence Take into account the different rates of change at different positions in a gene, codon, and among different nucleotides. For example, transitions are usually more likely than are transversions. Mathematical models can be used to compute how a tree might evolve. Maximum Likelihood Method Maximum likelihood method will identify the tree that most likely produced the observed data. They incorporate more information about evolutionary change than do parsimony methods. They are easier to treat in a statistical framework. The disadvantages are that they are computationally intensive and require explicit models of evolutionary change. Testing the accuracy of phylogenetic methods Biologists conducted experiments in living organisms and computer simulations that have demonstrated the effectiveness and accuracy of phylogenetic methods. Experiment: used viral culture of bacteriophage T7 to test accuracy. Phylogenetic methods reconstructed the known history correctly and were sensitive to highly variable rates of evolutionary change. Concept 16.3: Phylogeny Makes Concept 16.3: Phylogeny Makes Biology Comparative and Predictive Phylogeny can clarify the origin and evolution of traits in many biological processes Self-compatibility evolved three times in the angiosperm genus Leptosiphon from selfincompatibility, eliminating dependence on outside pollinators and accompanied by reduced petal size Zoonotic diseases are caused by infectious organisms transmitted from an infected animal of a different species, which can be traced through phylogenetic analysis; HIV-1 from chimpanzees and HIV-2 from sooty mangabeys Phylogenies can reconstruct the morphology, behavior, or nucleotide and amino acid sequences of ancestral species and allow for the understanding of complex traits Molecular clocks can be used to predict evolutionary divergence times using genes that evolve at a constant rate, calibrated using independent data such as the fossil record and biogeographic dates HIV-1 was present in human populations in Africa for at least 50 years before its emergence as a global pandemic, and immunodeficiency viruses have been transmitted repeatedly into human populations from multiple primates for more than a century Concept 16.4: Phylogeny Is the Basis of Biological Classification Binomial Nomenclature Developed by Carolus Linnaeus Each species has two names - the specific name and the group of closely related species (genus) Genus name is capitalized, specific name is lowercase and both are italicized Species and genera are further grouped into hierarchical system of higher taxonomic categories Taxon above genus is family Family names based on name of a member genus There are nothing that makes a family in one group equivalent to a "family" in another group Taxonomy Any group of organisms treated as unit in a biological classification system is called a taxon Families are grouped into orders, orders into classes, and classes into phyla (singular phylum), and phyla into kingdoms Ranking of taxa within Linnaean classification is subjective Biologists today recognize the tree of life as the basis for biological classification Triphyletic and paraphyletic groups are inappropriate as taxonomic units because they do not correctly reflect evolutionary history Unique Scientific Names Rules of biological nomenclature are designed so that there is only one correct scientific name for any single recognized taxon and a given scientific name applies only to a single taxon Sometimes the same species is named more than once, but the valid name is the first name that was proposed Using Phylogenetics to Study Evolutionary Relationships Monophyletic group is a historical group of related species, or a complete branch on the tree of life A true monophyletic group can be removed from a phylogenetic tree by a single cut in the tree Virtually all taxonomists now agree that polyphyletic and paraphyletic groups are inappropriate as taxonomic units because they do not correctly reflect evolutionary history There are many monophyletic groups on any phylogenetic tree Resurrecting Protein Sequences from Extinct Organisms in the Laboratory Sequences of many ancient genes and proteins can be reconstructed if enough information is available about the genomes of their descendants Biologists have reconstructed gene sequences from many species that have been extinct for millions of years Detailed mathematical models are used to reconstruct protein sequences from species that have been extinct for millions or even billions of years