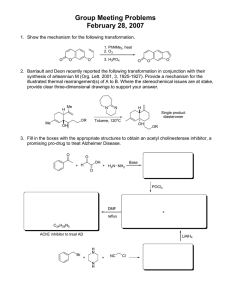
Heterocyclic Chemistry in Kinase Targeted Drug Discovery Kazuya Kano, Yoshiaki Washio, Yasushi Miyazaki, Yutaka Maeda, Hideyuki Sato, Masato Nakano, Masaichi Hasegawa Naohiko Nishigaki, Jun Tang GlaxoSmithKline K. K., Tsukuba Research Laboratories, Chemistry Department Introduction Where the Selectivity of this Series Comes from? キナーゼ: ATPを介して基質であるセリン/スレオニンもしくはチロシ ンの水酸基をリン酸化する酵素のことである。 Phe 80 Regulates: activity, location, degradation, conformation. Many implicated in disease - especially cancer... Kinase Systems Based Research to identify a molecule for a kinase (我々はSystems Based Researchと呼 ばれる手法に基づいて創薬研究を行っている。この手法は従来の疾患由 来の創薬研究とは異なる) •Start with a target family, not disease •Allows us to apply what we learn from one member of the class to others of interest •More efficient - not a fresh start with each new target •Build and use a knowledge base Why it works •ATP site - conserved but not optimized for ATP •Large database of structures allows for: •greater understanding of key pharmacophores and SAR •improved homology models •novel template design Lys 85 CDK 2 Gatekeeper GSK-3 N N H Br HN Lysine area H2N Biphenylureas, called “induced fit” moiety, at 5-position greatly improve potency against angiogenesis related kinases such as VEGFR’s, Tie2 and EphB4. GW 832757X shows great Cellular activity(4.5nM) Phe 80 CDK 2 Gatekeeper Leu 132 As Angiogenesis Inhibitor Leu 132 H N Imported Data From Chemically Aware Spreadsheet O Lysine area Lys 85 CF3 Lead Example 2 H N H2N H N 6.57 5.23 N H 4.96 6.81 6.39 HN 8.94 8.63 8.57 F N H CF3 8.63 Ar N R Gate Keeper N H N N Gate Keeper induced fit O N Cl N H N H N HN Pan kinases <= 6.1 induced fit inner hydrophobic OH Lys N Glu H H O H N H Scaffold N Lys area O O O O H N Asp Outer R Hydrophobic O N N H O N O H N H inner hydrophobic N H Scaffold O N Outer H Hydrophobic Solvent Front GW 850576X Rock1 pIC50 = 7.2 JNK3 pIC50 = 6.9 VEGFR2, C-Fms pIC50 < 6.0 O Lys N Glu H H O H Lys area O O O R HN Sugar Pocket N O Asp OH Sugar Pocket Solvent Front In other cases X-ray structure of ‘697X with c-fms In C-Fms GSK3 Inhibitor Design-starting from Tie2 legacy-Example 4 S O GW 671819X GSK3 pIC50 = 7.1 CDK2 pIC50 = 4.7 H N HN N N O S NH2 O N H GW 822547X CDK2 pIC50 = 9.5 Super selectiv e CDK2 inhibitor Val 135 N Pro N H H Tyr NN NNH O H N Lys N O NH2 N O O HO O O HN Asp N GW-671819X SB-591475-A N H N H O O N SB-591475-A and GW-671819X docked into GSK-3 SB-591475A GSK3 pIC50 = 7.5 GW 833547X CDK2 pIC50 = 9.5 CDK1 pIC50 = 6.3 N N O Strategy of GSK3 Inhibitor Pursuance New Template Development-Example 3 GW 680665X N F Br Core Structure C-FMS VEGFR2 TrJNK3 GSK3 6.23 6.10 5.82 5.87 OH Gate Keeper pIC50 induced fit VEGFR2 src others N inner hydrophobic OH Lys N Glu H H O H N H Scaffold N Lys area O O O O H N Asp Outer R Hydrophobic N O H Gate Keeper induced fit inner hydrophobic Lys N Glu H O H Lys area O O O H O N Outer Hydrophobic Sugar Pocket H N 136 NH2 New Template Development-Example 1 H N Asp NH2 Overlap of GW 818954X and GW 833547X H N 133 O O N HN N N H N GW 828604X GSK3 pIC50 = 8.3 L6 glycogen Accumulatioin Cellular Assay pEC50 = 6.4 O N GW 818954X CDK2 pIC50 = 8.6 YAK1 pIC50 = 8.2 CDK1 pIC50 = 7.9 Publication Example 1 H2N N N O H S N O N H O O N 20 Months O NH2 N N N H GW 847320X CDK2 pIC50 = 9 Cellular Assay IC50 = 3-5 uM Lead Example 4 NH N H S O CF3 O N HN N H2N GW 832757X VEGFR2 pIC50 = 8.5 TIE2 pIC50 = 8.9 50 nM Cellular Activ ity(Huv ec V) TRL Chemists SBR(5410) N F O N N 4.59 N Lead Example 3 Scaffold 1 7.3 In the case of c-fms, isoquinoline demonstrates unique SAR caused by FLIP of the template. This binding mode is supported by x-ray structure analysis. O H2N S O GW 847320X CDK2 pIC50 = 9.0 Yak1 pIC50 = 8.3 HN 7.21 As C-FMS Inhibitor Aminopyrazole versus Pyrrolopyrimidine Similarity and Difference H HN N TRL Examples of Successful Kinase Inhibitor Heterocycles N H O H N H N SB 736764 c-Fms pIC50 = 7.67 VEGFR2 pIC50 = 6.72 7 O GW 832757X F N 6.4 CF3 Pyrrolopyrimidine Hit Series-Example 2 HN 5 EPHB4 F H N H NH 2 O VEGFR 3 N CF3 H N Lys 33 Based on our research, some new information have been added to kinase system including the concept of Lysine Area and how to design a selective CDK-2 inhibitor. H N N H GW 792416X GW 824374X O H N HN VEGF R2 TIE-2 F R H2N H N 2 Lys 33 N O O H N S Ar PIC50 Ar GW 824372X N H2N 09 October 2002 R GSK-3 H N 5.5 5.5 <4.8 GW 586002X The best in class N H O 2 4 H NH N HO HN Attach acyl- to improv e the interaction w ith FLP and profile Cut off 3-position substituent Solvent Front O N N O N Optimization of 2-position substituent Front lipophilic pocket Sugar Pocket Salt bridge O 3 O Asp NH2 N O GW 828604X GSK3 pIC50 = 8.33 CDK2 pIC50 = 6.34 Solvent Front The Difference between this Hit and Historical Inhibitor O O S NH H N N O NH O O S NH2 Modeling Analysis Comparison As ALK5 Inhibitor TGFb-Alk5 program for renal fibrosis needs new back-up series because of selectivity issue over Alk4. Hit series shown here shows excellent selectivity versus ALK4. Before modification N After modification 1 H N N R NH R S pIC50 ALK4 ALK5 Ar R Br GW 278534X GW 798644X CDK-2(pic50): 8.2 GSK-3(pic50): 7.3 CDK-2(pic50): 7.6 GSK-3(pic50): 5.6 pIC50 ALK4 ALK5 Ar HN HN N NH 4.61 7.71 NH2 6.3 7.08 6.93 7.13 6.2 6.61 N OH OH S NH 4.61 7.28 NH2 N HN OH Overlap of X-ray structures of GW 278534X and GW 798644X in the same CDK-2 protein. Ar 5 NH 4.61 CF3 7.15 H Overlap of GW 671819X and SB-591475A Overlap of GW 809653X and SB-591475A Printed by Information Design