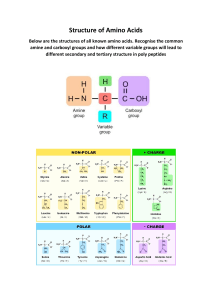
Name _______________ ID _____________ Chem 153A – Homework 2 due 1/24 @ 11:59 pm 1. Consider the below reactions: Glucose + fructose ↔ sucrose ATP + H2O ↔ ADP + Pi ΔG = +27 kJ/mol ΔG = -30 kJ/mol a. Draw reaction coordinate diagrams for both of the above reactions. b. Assume the above reactions have been coupled. What is the final overall reaction and what is its ΔG? Is it spontaneous? 2. The equilibrium constant for the below reaction is 0.09: R→P a. Assuming the reaction has reached equilibrium, if [P] = 0.05 mM, what is [R]? b. If [R] = 0.2 mM, what direction would you expect the reaction to proceed? Name _______________ ID _____________ 3. Consider the relationship between ΔG° and Keq. a. Write out the equation that most directly summarizes this relationship. b. If K > 1, what does this mean for the value of ΔG°? What does this mean for the ratio of products to reactants? What about when K < 1? 4. Explain the difference between equilibrium and steady state. 5. This image below shows a group of fatty acids being solvated by water. a. Describe the shell of water surrounding each fatty acid. Is this arrangement thermodynamically favorable? b. What will happen to these fatty acids? Describe the likely physical process and give any relevant thermodynamic details. c. How does your insights from part a and part b relate to protein folding? Name _______________ ID _____________ 6. An experiment was performed on four samples of RNase A (an enzyme that catalyzes the degradation of RNA), garnering the following results: Sample details: - 100 μL of reaction buffer was added to Sample 1. - 100 μL of reaction buffer + urea was added to Sample 2. - 100 μL of reaction buffer + urea + 2-mercaptoethanol was added to Sample 3. - 100 μL of reaction buffer + urea + 2-mercaptoethanol was added to Sample 4. The sample was then dialyzed to remove the urea and 2-mercaptoethanol. - All four samples were then tested for RNase activity through adding a small RNA oligonucleotide with a fluorescence dye and quencher (when the RNA is degraded, the dye is freed, creating fluorescence). a. What does this experiment demonstrate about proteins and protein folding? b. What happens to the RNase A in Sample 3 that does not occur in Sample 2? Provide physical details. Name _______________ ID _____________ 7. The following two diagrams are energy landscapes representing two distinct hypotheses relating to protein folding. a. What does “N” refer to? Why is it meaningful that N is (spatially) lower in each energy diagram? b. What is the first diagram “saying” about protein folding? What issue does it raise? Describe this issue. c. How does the second diagram address the first diagram? d. Evaluate these hypotheses – are they correct? Why or why not? Name _______________ ID _____________ 8. The below diagram shows a polypeptide folding into a protein. a. What category of amino acid residues do the black dots represent? b. The spontaneous folding of proteins occurs due to the net action of several different factors. Describe these factors physically and thermodynamically. Makes sure to include whether these factors work against or in favor of spontaneous folding. c. Draw a free-energy landscape to go along with your explanation in part b. In it, include the unfolded state, the molten globule state, and the native (fully folded) state. Name _______________ ID _____________ 9. The representation below shows a common secondary structure. a. What do the red and blue refer to in the diagram above? b. What is the primary interaction that stabilizes this structure? c. What spatial pattern is formed by the R groups in each peptide chain? d. Is this secondary structure parallel or antiparallel? Describe two ways you know this to be true. e. Given your answer to part d, how would these peptide chains be connected to one another? Describe and draw your answer (molecular details not requisite). Name _______________ ID _____________ 10. The representation to the right is part of a protein structure. a. What level of protein structure does this represent? What is the name of this particular structure? b. What non-covalent interaction stabilizes this structure (primarily)? Draw three of these interactions on the representation with dipoles clearly labeled. c. Indicate the position of three amino acid side chains in the structure (draw an R for each side chain). d. This structure has an odd shape overall. What word do we usually use to describe the bend in this structure? Which two amino acids could be responsible for this “bend” and why do they have this impact? e. A frameshift mutation changes several amino acids in the structure, causing there to be several Arg residues in a row. What’s the impact of this mutation on this structure? Name _______________ ID _____________ 11. Use the Ramachandran plot below to answer the following questions. a. What do φ and ψ represent? Use a drawing in your explanation. b. What does a Ramachandran plot represent? c. Label the three heavily populated areas on the chart. What are they? d. Why are there completely blank (white) spaces on a Ramachandran plot? What is/isn’t happening there and why? Name _______________ ID _____________ 12. For the following levels of protein structure, briefly describe the forces (intramolecular or intermolecular) involved in these structures. a. Primary structure b. Secondary structure c. Tertiary structure d. Quaternary structure 13. Protein secondary structures can be amphipathic. a. Draw an example of an amphipathic secondary structure using relevant labels to communicate your understanding. b. Briefly explain how each kind of amphipathic secondary structure can arise from the primary sequence (hint: what pattern do we see in the primary sequences of each?). c. What structural function could an amphipathic secondary structure serve? Provide two possibilities.