06/07/2022, 09:43
SUPER-FOCUS | workshop2022
FAME Metagenomics workshop 2022
Home
View On
GitHub
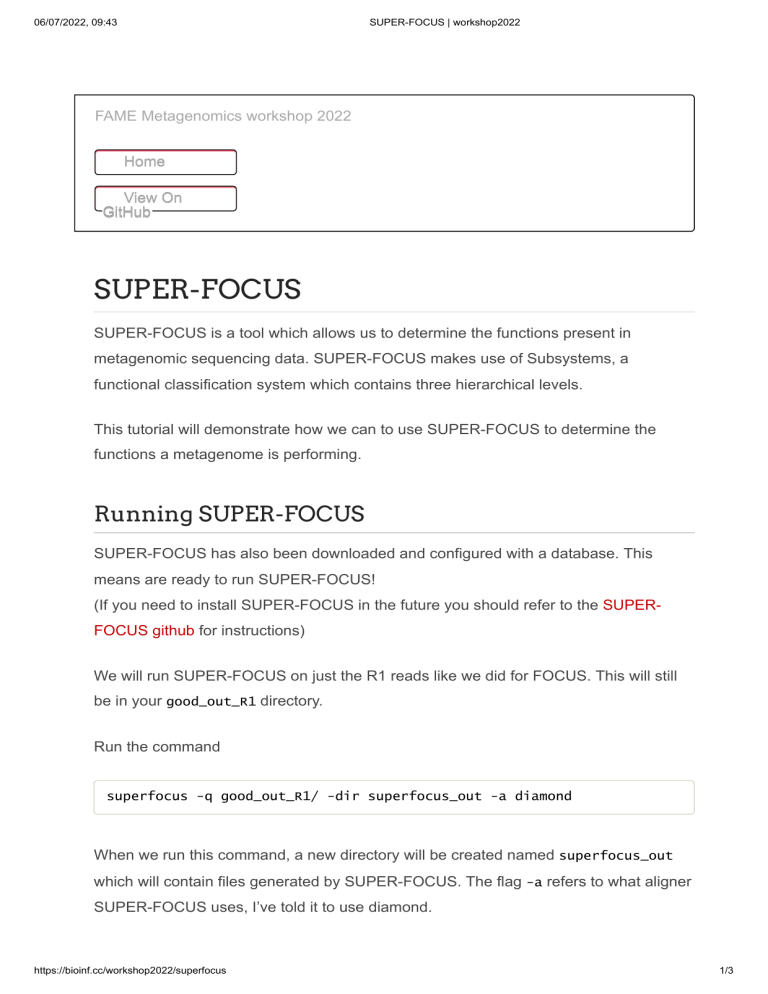
SUPER-FOCUS
SUPER-FOCUS is a tool which allows us to determine the functions present in
metagenomic sequencing data. SUPER-FOCUS makes use of Subsystems, a
functional classification system which contains three hierarchical levels.
This tutorial will demonstrate how we can to use SUPER-FOCUS to determine the
functions a metagenome is performing.
Running SUPER-FOCUS
SUPER-FOCUS has also been downloaded and configured with a database. This
means are ready to run SUPER-FOCUS!
(If you need to install SUPER-FOCUS in the future you should refer to the SUPERFOCUS github for instructions)
We will run SUPER-FOCUS on just the R1 reads like we did for FOCUS. This will still
be in your good_out_R1 directory.
Run the command
superfocus -q good_out_R1/ -dir superfocus_out -a diamond
When we run this command, a new directory will be created named superfocus_out
which will contain files generated by SUPER-FOCUS. The flag -a refers to what aligner
SUPER-FOCUS uses, I’ve told it to use diamond.
https://bioinf.cc/workshop2022/superfocus
1/3
06/07/2022, 09:43
SUPER-FOCUS | workshop2022
Great, now what?
We can start to look at the output which SUPER-FOCUS by taking looking in the output
directory
cd superfocus_out
You’ll notice a few files ending with .m8. These are alignment files generated by
superfocus.
More importantly, you should notice the files, output_subsystem_level_1.xls
output_subsystem_level_1.xls output_subsystem_level_1.xls . Each of these
files provides details on the prevalence of each function belonging to the corresponding
level. All three levels are contained in the file output_all_levels_and_function.xls
To look at the level 1 output run the command
column -t -s $'\t' -n string output_subsystem_level_1.xls
| less
Here the first four columns correspond to the normalised read counts of each sample,
and the second four columns contain the percent abundance of each function.
(Note that the read counts in the superfocus output have been normalised and this is
why read counts have decimal values. if you would prefer to have the raw, unnormalised read counts in the output, make sure to run SUPER-FOCUS with the flag -n
0)
When you are done looking at the output, press the letter ‘q’ on your keyboard.
Visualising SUPER-FOCUS with Krona
We can build a Krona plot on our SUPER-FOCUS output just like we did for our Kraken
output earlier today.
Again, we need to rearrange the output into a format which Krona can understand. We
can rearrange it using this bash command.
https://bioinf.cc/workshop2022/superfocus
2/3
06/07/2022, 09:43
SUPER-FOCUS | workshop2022
tail -n +5 output_all_levels_and_function.xls | awk -F '\t' '{n=$4+$5+$6
This creates a file, superfocus_out_krona.tsv which can be read by Krona.
Next we can generate our krona plot by running the command
ktImportText superfocus_out_krona.tsv -o superfocusKronaPlot.html
Next download the Krona html file to your desktop using WinSCP. You can open this
html file in your favourite browser to reveal a plot of the distribution of functions in the
samples. You can zoom in and zoom out to see the different levels of annotations.
scp -r grig0076@115.146.84.253:/home/grig0076/superfocus_out/superfocus_
Congratulations on making it to the end of the tutorial! I hope you enjoyed it
Feeling lazy? Here’s the final product!
https://bioinf.cc/workshop2022/superfocus
3/3