Chemistry of Biological and Natural Products
Experiment 1
Objectives: To determine the available nitrogen in the soil sample by Kjeldahl Method.
Theory:
A soil analysis is a process by which elements such as P, K, Ca, Mg, Na, S, Mn, Cu and Zn are
chemically extracted from the soil and measured for their “plant available” content within the soil
sample.
Significance of Soil Analysis:
It increases the knowledge of what nutrients are especially available in our soil.
It reduces the environmental impacts due to soil amendments.
It increases the efficiency of resource inputs such as fertilisers and water.
It helps to predict the nutritional values needed for crop production.
It helps to evaluate the fertility status of soils of a country or a state or a district.
Procedure for Taking Good Soil Samples:
Determine the soil unit (or plot).
Make a traverse over the soil unit (or plot).
Clean the site (with spade) from where soil sample is to be collected.
Insert the spade into soil.
Standing on the opposite side, again insert the spade into soil.
A lump of soil is removed.
A pit of ‘V’ shape is formed. Its depth should be 0-6" or 0-9" or 0-12" (i.e., Depth of
tillage).
Take out the soil-slice (like a bread slice) of ½ inch thick from both the exposed surface of
the pit from top to bottom. This slice is also termed furrow-slice. To collect the soil-slice
spade may be used. Collect the soil samples in a polyethylene bucket.
Collect furrow-slices from 8-10 or sometimes 20-30 sites. Select the sites at random in a
zigzag (or criss-cross) manner. Distribute the sites throughout the entire soil unit (plot). In
lieu of spade auger may be used. Do not take the prohibited samples and local problem
soils.
Furnish the following information in two sheets of thick paper with the sample. One sheet
is folded and kept inside the bag. Another sheet is folded and attached to the bag.
Available Nitrogen Content in Soil:
Nitrogen is one of the major elements required for life. It will stimulate above ground growth, and
produces the rich green colour that is the characteristic of healthy plants, because of this Nitrogen
is essential for plant life. 78% of the atmosphere is covered by molecular Nitrogen (N2); this form
of Nitrogen cannot be used by animals. This molecular Nitrogen must first combine with Oxygen
or Hydrogen to produce compounds such as Ammonia or Nitrate, or some other organic form of
Nitrogen. This is called Nitrogen Fixation. Some Nitrogen Fixation occurs by lightning and some
other by blue green algae. However, the bulk of Nitrogen Fixation is preferred by bacteria living
in the soil. Some of the Nitrogen Fixation bacteria were living free in the soil, while the others
were living within the root nodules of some plants such as soya bean, peanut, beans, clover, alfalfa,
etc. Because of Ammonia or Ammonium is produced by the decomposition process, the
decomposition of materials in the forest is also a source of Nitrogen. The movement of Nitrogen
from the atmosphere into inorganic forms, followed by the incorporation of Nitrogen into plant
matter is represented as the Nitrogen Cycle, which is shown in the figure given below.
The rate of plant growth is proportional to the rate of nitrogen supply. If the soil is deficient in
Nitrogen, the plants become stunted and pale. However, an excess of Nitrogen can damage the
plants just as over-fertilizing the lawn can burn and damage the grass.
Apparatus:
Kjeldahl Digestion Assembly, Ammonia Distillation Assembly.
Principle:
The Kjeldahl method permits the available nitrogen to be precisely determined in the plant and in
the soil. The method of determination involves three successive phases which are,
1. Digestion:
Digestion of the organic material is carried out by digesting the sample with Con. H 2SO4 in the
presence of CuSO4.H2O as a catalyst and K2SO4 which raise the digestion temperature. The
organic material decomposes into several components i.e.,
C → CO2, O → H2O and N → NH3
In the organic matter, some nitrates are present, most of which are lost during the digestion. The
loss may be disregarded for most soils. Since the amount of NO3- - N is far lesser than the Organic
Nitrogen.
2 C6H3 (OH) NH2COO + 26 H2SO4 → (NH4) 2SO4 + 25 SO2 + 14 CO2 + 28H2O
2. Distillation:
The Ammonia content of the digest is determined by distillation with excess NaOH and absorption
of the evolved NH3 is in standard HCl.
(NH4)2SO4 + 2 NaOH → Na2SO4 + 2 NH3 + 2 H2O
NH3 +HCl → NH4Cl
3. Volumetric Analysis:
The excess of standard HCl is titrated against standard NaOH using Methyl Red as an indicator.
The decrease in the multi equivalence of acid as determined by acid-base titration, which gives a
measure of the N content of the sample. The end point is determined by a change of colour from
pink to yellow.
2 HCl + 2 NaOH → 2 NaCl + H2O
Significance:
The chemical analysis of the soil for nitrogen is less precise when the requirement for this element
needs to be forecast over a longer period of time, as they vary not only with species, but with the
phase of growth and season as well. Therefore, the chemical test for NO3- and NH4+ signifies the
momentary status when the sample is taken and measures must be taken instantaneously. The
analysis of the extractable Nitrogen content of the soil using a given extractable method.
In reaction to crop response study provides a basis of Nitrogen fertility levels, which will
rationalize the use efficiency of Nitrogen fertilizer content of the soil are also needed for the
evaluation of C-N ratios of soils which give an indication of the process of transformation of
organic Nitrogen to available Nitrogen like ammoniated nitrate Nitrogen.
Observations and Calculations:
Results: The % of N in the given soil sample is 0.0672%.
Experiment 2
Objective:
To determine chemical parameters such as hardness, alkalinity, and chemical oxygen demand
COD) of water samples.
Theory:
It is needless to emphasize the importance of water in our life. Without water, there is no life on
our planet. We need water for different purposes. We need water for drinking, for industries, for
irrigation, for swimming and fishing, etc.
Water for different purposes has its own requirements for composition and purity. Each body of
water needs to be analysed on a regular basis to confirm to suitability. The types of analysis could
vary from simple field testing for a single analyte to laboratory based multi-component
instrumental analysis. The measurement of water quality is a very exacting and time-consuming
process, and a large number of quantitative analytical methods are used for this purpose.
1. Total hardness:
Theory:
Hardness in water is that characteristic, which “prevents the lathering of soap”. This is due to
presence in water of certain salts of calcium, magnesium and other heavy metals dissolved in it. A
sample of hard water, when treated with soap does not produce lather, but on other hand forms a
white scum or precipitate. This precipitate is formed, due to the formation of insoluble soaps of
calcium and magnesium.
Thus, water which does not produce lather with soap solution readily, but forms a white curd, is
called hard water. On the other hand, water which lathers easily on shaking with soap solution, is
called soft water. Such water consequently does not contain dissolved calcium and magnesium
salts in it.
Temporary or carbonate hardness: It is caused by the presence of dissolved bicarbonates of
calcium, magnesium and other heavy metals and the carbonate of iron. Temporary hardness is
mostly destroyed by mere boiling of water, when bicarbonates are decomposed, will produce
insoluble carbonates or hydroxides, which are deposited as a crust at the bottom of vessel.
Permanent or non-carbonate hardness: It is due to the presence of chlorides and sulphates of
calcium, magnesium, iron, and other heavy metals. Unlike temporary hardness, permanent
hardness is not destroyed on boiling.
The degree of hardness of drinking water has been classified in terms of the equivalent
CaCO3 concentration as follows:
Soft
Medium
Hard
Very Hard
0-60mg/L
60-120mg/L
120-180mg/L
>180mg/L
In a hard water sample, the total hardness can be determined by titrating the Ca2+ and Mg2+ present
in an aliquot of the sample with Na2EDTA solution, using NH4Cl-NH4OH buffer solution of pH
10 and Eriochrome Black-T as the metal indicator.
Na2H2Y (Disodium EDTA solution) → 2Na+ + H2YMg2+ + HD2- (blue) → MgD (wine red) + H+
D (metal-indicator complex, wine red colour) + H2Y- →Y- (metal EDTA complex colourless) +
HD- (blue colour) + H+
Ethylenediamine tetra-acetic acid (EDTA) and its sodium salts form a chelated soluble complex
when added to a solution of certain metal cations. If a small amount of a dye such as Eriochrome
black T is added to an aqueous solution containing calcium and magnesium ions at a pH of 10 ±
0.1, the solution will become wine red. If EDTA is then added as a titrant, the calcium and
magnesium will be complexed. After sufficient EDTA has been added to complex all the
magnesium and calcium, the solution will turn from wine red to blue. This is the end point of the
titration.
Units of Hardness:
1. Parts per million (ppm): Is the parts of calcium carbonate equivalent hardness per 106 parts of
water, i.e, 1 ppm = 1 part of CaCO3 eq hardness in 106 parts of water.
2. Milligram per litre (mg/L): Is the number of milligrams of CaCO3 equivalent hardness present
per litre of water. Thus:
1 mg/L = 1 mg of CaCO3 eq hardness per L of water.
3. Clarke’s degree (oCl): Is number of grains (1/7000 lb) of CaCO3 equivalent hardness per gallon
(10 lb) of water. Or it is parts of CaCO3equivalent hardness per 70,000 parts of water. Thus,
1oClarke = 1 grain of CaCO3 eq hardness per gallon of water.
4. Degree French (oFr): Is the parts of CaCO3 equivalent hardness per 105 parts of water. Thus,
1o Fr = 1 part of CaCO3 hardness eq per 105 parts of water.
Relationship Between Various Units of Hardness:
1ppm =1 mg/L=0.1oFr =0.07oCl
1mg/L=1 ppm=0.1oFr =0.07oCl
1oCl=1.43oFr=14.3 ppm=0.7omg/L
1oFr=10 ppm=10 mg/L=0.7oCl
2. Alkalinity:
Theory:
Alkalinity is an aggregate property of the water sample which measures the acid-neutralizing
capacity of a water sample. It can be interpreted in terms specific substances only when a complete
chemical composition of the sample is also performed. The alkalinity of surface water is due to
the carbonate, bicarbonate and hydroxide content and is often interpreted in terms of the
concentrations of these constituents. Higher the alkalinity, greater is the capacity of water to
neutralize acids. Conversely, the lower the alkalinity, the lesser will be the neutralizing capacity.
Alkalinity of sample can be estimated by titration with standard H2SO4 or HCI solution. Titration
to pH 8.3 or decolourisation of phenolphthalein indicator will indicate complete neutralization of
OH- and 1/2 of CO32-, while to pH 4.5 or sharp change from yellow to orange of methyl orange
indicator will indicate total alkalinity.
To detect the different types of alkalinity, the water is tested for phenolphthalein and total
alkalinity, using Equations:
Where,
A = titrant (mL) used to titrate to pH 8.3
B = titrant (mL) used to titrate to pH 4.5
N = normality of the acid (0.02N H2SO4 for this alkalinity test)
50,000 = a conversion factor to change the normality into units of CaCO3
Once PA and TA are determined, then three types of alkalinities, i.e, hydroxides, carbonates and
bicarbonates can be easily calculated from the table:
Result of Titration OH alkalinity as CaCO3 CO3 alkalinity as CaCO3 HCO3 alkalinity as CaCO3
PA = 0
0
0
TA
PA < 1/2TA
0
2PA
TA - 2PA
PA = 1/2TA
0
2PA
0
PA > 1/2TA
2PA - TA
2(TA - PA)
0
PA = TA
TA
0
0
3. Chemical Oxygen Demand (COD):
Theory:
COD is used as a measure of oxygen equivalent to organic matter content of a sample that is
susceptible to oxidation by a strong chemical oxidant. For samples from a specific source, COD
can be related empirically to BOD. COD determination has advantage over BOD determination in
that the result can be obtained in about 5 hours as compared to 5 days required for BOD test.
The organic matter gets oxidized completely by K2Cr2O7 in the presence of H2SO4 to produce
CO2 and H2O. The excess of K2Cr2O7 remained after the reaction is titrated with ferrous
ammonium sulphate. The dichromate consumed gives the O2 required for oxidation of organic
matter.
DRINKING WATER QUALITY STANDARDS:
Sl. No.
1
2
3
4
Characteristic/Parameter
Colour
Odour
Turbidity
pH
BIS
ICMR
WHO
5
Agreeable
10 NTU
6.5-8.5
2.5
Unobjectionable
5 NTU
7.0-85
Unobjectionable
2.5 NTU
7.0-8.5
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
TDS
Hardness
Ca
Mg
CL
Sulphate
Fe
Nitrate
Phenolic compounds
Cd, Sc
Cu, As
Cyanides
Pb
Anionic detergents
PAH
Residual Chlorine
Pesticides
BIS- Beaurau of Indian Standards.
ICMR- Indian Council for Medical Research.
WHO- World Health Organization
Observations:
Titrant used: 2.6 mL
500 mgl
300 mgl
75 mgl
30 mgl
250 mgl
200 mgl
0.3 mgl
45 mgl
0.001 mgl
0.01 mgl
0.05 mgl
0.05 mgl
0.1 mgl
0.2 mgl
0.2 mgl
Absent
500 mgl
300 mgl
75 mgl
50 mgl
200 mgl
200 mgl
0.1 mgl
20 mgl
0.001 mgl
0.05 mgl
-
500 mgl
200 mgl
75 mgl
30 mgl
200 mgl
200 mgl
0.1 mgl
45 mgl
0.001 mgl
0.01 mgl
0.01 mgl
0.01 mgl
0.01 mgl
-
Titrant used: 1.6 mL
Titrant used: 6.0 mL
Calculations:
1. Well water
Volume of EDTA used= 2.6 mL
Molarity of EDTA = 0.01 M
Volume of the water sample = 10 mL
Therefore, the total hardness of the sample is
=
= 260 ppm
2. Tap water
Volume of EDTA used= 1.6 mL
Molarity of EDTA = 0.01 M
Volume of the water sample = 10 mL
Therefore, the total hardness of the sample is
=
= 160 ppm
3. Sea water
Volume of EDTA used= 6.0 mL
Molarity of EDTA = 0.01 M
Volume of the water sample = 10 mL
Therefore, the total hardness of the sample is
=
Results:
Hardness of well water is 260 ppm.
Hardness of tap water is 160 ppm.
Hardness of sea water is 600 ppm.
= 600 ppm
Experiment 3
Objective:
To Estimate the Saponification value of oils
Theory:
Fats and oils are the principle stored forms of energy in many organisms. They are highly reduced
compounds and are derivatives of fatty acids. Fatty acids are carboxylic acids with hydrocarbon chains of
4 to 36 carbons, they can be saturated or unsaturated. The simplest lipids constructed from fatty acids
are triacylglycerols or triglycerides. Triacylglycerols are composed of three fatty acids each in ester
linkage with a single glycerol. Since the polar hydroxyls of glycerol and the polar carboxylates of the
fatty acids are bound in ester linkages, triacyl glycerols are non-polar, hydrophobic molecules, which are
insoluble in water.
Saponification is the hydrolysis
of fats or oils under basic
conditions to afford glycerol and
the salt of the corresponding
fatty acid. Saponification
literally means "soap making".
It is important to the industrial
user to know the amount of free
fatty acid present, since this determines in large measure the refining loss. The amount of free fatty acid is
estimated by determining the quantity of alkali that must be added to the fat to render it neutral. This is
done by warming a known amount of the fat with strong aqueous caustic soda solution, which converts
the free fatty acid into soap. This soap is then removed and the amount of fat remaining is then
determined. The loss is estimated by subtracting this amount from the amount of fat originally taken for
the test.
The saponification number is the number of milligrams of potassium hydroxide required to neutralize the
fatty acids resulting from the complete hydrolysis of 1g of fat. It gives information concerning the
character of the fatty acids of the fat- the longer the carbon chain, the less acid is liberated per gram of fat
hydrolysed. It is also considered as a measure of the average molecular weight (or chain length) of all the
fatty acids present. The long chain fatty acids found in fats have low saponification value because they
have a relatively fewer number of carboxylic functional groups per unit mass of the fat and therefore high
molecular weight.
Principle:
Fats (triglycerides) upon alkaline hydrolysis (either with KOH or NaOH ) yield glycerol and potassium or
sodium salts of fatty acids (soap) .
Materials required:
1) Fats and Oils [coconut oil, sunflower oil]
2) Conical Flask
3) 100ml beaker
4) Weigh Balance
5) Dropper
6) Reflux condenser
7) Boiling Water bath
8) Glass pipette (25ml)
9) Burette
Reagents required:
1)
2)
3)
4)
5)
Ethanolic KOH(95% ethanol, v/v)
Potassium hydroxide [0.5N]
Fat solvent
Hydrochloric acid[0.5N]
Phenolphthalein indicator
Procedure:
1)
Weigh 1g of fat in a tared beaker and dissolve in about 3ml of the fat solvent [ ethanol /ether
mixture].
2) Quantitatively transfer the contents of the beaker three times with a further 7ml of the solvent.
3) Add 25ml of 0.5N alcoholic KOH and mix well, attach this to a reflux condenser.
4) Set up another reflux condenser as the blank with all other reagents present except the fat.
5) Place both the flasks in a boiling water bath for 30 minutes .
6) Cool the flasks to room temperature .
7) Now add phenolphthalein indicator to both the flasks and titrate with 0.5N HCl .
8) Note down the endpoint of blank and test .
9) The difference between the blank and test reading gives the number of millilitres of 0.5N KOH
required to saponify 1g of fat.
10) Calculate the saponification value using the formula :
Saponification value or number of fat = mg of KOH consumed by 1g of fat.
Weight of KOH = Normality of KOH * Equivalent weight* volume of KOH in litres
Volume of KOH consumed by 1g fat = [Blank – test]ml
Procedure:
1
.
2
.
3
.
4
.
5
6
7
8
Mixtureethanol+KOH
9
10
0
11
0
(AFTER ADDING THE
INDICATOR)
12
0
13
0
REPEAT THE SAME WITH THE BLANK AND NOTE THE END POINTS.
Observations:
1. Oil/fat used- Coconut oilBLANK
READING-20.00 mL
TEST
READING-11.00 mL
CalculationsTITLE
INITIAL BURETTE
FINAL BURETTE
VOLUME OF HCL
READING
READING
RUN DOWN
TEST
0.0
11.00
11.00
BLANK
0.0
20.00
20.00
VOLUME OF KOH CONSUMED BY 1g FAT= (20.0-11.0) = 9.00 Ml
WEIGHT OF KOH= NORMALITY OF KOH*EQUIVALENT WEIGHT*VOLUME OF KOH IN L
0.5*56*9/1000=0.252g=252 mg=saponification number of coconut oil
2. Oil/fat used- Butter-
READINGS- 12.6 mL
Calculations:
TITLE
TEST
BLANK
INITIAL BURETTE
READING
0.0
0.0
FINAL BURETTE
READING
12.60
20.00
VOLUME OF HCL
RUN DOWN
12.60
20.00
VOLUME OF KOH CONSUMED BY 1g FAT= (20.0-12.6) = 7.4 Ml
WEIGHT OF KOH= NORMALITY OF KOH*EQUIVALENT WEIGHT*VOLUME OF KOH IN L
0.5*56*7.4/1000=0.2072=207.2 mg=saponification number of butter
Result:
The saponification number of coconut oil is 252 mg.
The saponification number of butter is 207.2 mg.
Experiment 4
Estimation of Iodine Value of Fats and Oils
Objective:
To determine the iodine value of fats and oils and thus estimate the unsaturation of the fats and
oils.
Theory:
Fats and oils are a mixture of triglycerids. Triglycerides are made up of three fatty acids linked
to glycerol by fatty acyl esters. Fatty acids are long chain hydrocarbons with carboxyl groups
(COOH groups). These fatty acids can be classified into saturated or unsaturated based on the
number of double bonds present in the fatty acid. Saturated fatty acids contain only single bond
between the carbon atoms and are tend to be solids at room temperature. Unsaturated fatty acids
contain double bonds between the carbon atom in addition to the single bonds present in the fatty
acid chain. They are likely to exists as liquids at room temperature. The double bonds present in
the naturally occurring unsaturated fats are in the Cis form. Trans fatty acids are associated with
health problems and cardiovascular diseases.
Unsaturated fatty acids can be converted into
saturated by the process of hydrogenation.
Depending upon the degree of unsaturation,
the fatty acids can combine with oxygen or
halogens to form saturated fatty acids. So it is
important to know the extend to which a fatty acid is unsaturated. There are different methods
for checking the unsaturation level in fatty acids, one among them is by determining the iodine
value of fats. Iodine value or number is the number of grams of iodine consumed by 100g of fat.
A higher iodine value indicates a higher degree of unsaturation.
unsaturated fatty acids
Principle:
Fatty acids react with a halogen [ iodine] resulting in the addition of the halogen at the C=C
double bond site. In this reaction, iodine monochloride reacts with the unsaturated bonds to
produce a di-halogenated single bond, of which one carbon has bound an atom of iodine.
After the reaction is complete, the amount of iodine that has reacted is determined by adding a
solution of potassium iodide to the reaction product.
ICl + KI ---------------> KCl + I2
This causes the remaining unreacted ICl to form molecular iodine. The liberated I2 is then
titrated with a standard solution of 0..1N sodium thiosulfate.
I2 + 2 Na2S2O3 -----------------> 2 NaI + Na2S2O4
Saturated fatty acids will not give the halogenation reaction. If the iodine number is between 070, it will be a fat and if the value exceeds 70 it is an oil. Starch is used as the indicator for this
reaction so that the liberated iodine will react with starch to give purple coloured product and
thus the endpoint can be observed.
Materials Required:
•
•
•
•
•
•
•
•
•
•
•
•
Iodine Monochloride Reagent
Potassium Iodide
Standardized 0.1 N Sodium thiosulphate
1% Starch indicator solution
Reagent bottle
Chloroform
Fat sample in chloroform
Iodination flask
Burette and burette stand with magnetic stirrer
Glass pipette
Measuring cylinder
Distilled water
Method:
1. Arrange all the reagent solutions prepared and the requirements on the table.
2. Pipette out 10ml of fat sample dissolved in chloroform to an iodination flask labeled as “TEST".
3. Add 20ml of Iodine Monochloride reagent in to the flask. Mix the contents in the flask
thoroughly.
4. Then the flask is allowed to stand for a half an hour incubation in dark.
5. Set up a BLANK in another iodination flask by adding 10ml Chloroform to the flask.
6. Add to the BLANK, 20ml of Iodine Monochloride reagent and mix the contents in the flask
thoroughly.
7. Incubate the BLANK in dark for 30 minutes.
8. Mean while, Take out the TEST from incubation after 30 minutes and add 10 ml of potassium
iodide solution into the flask.
9. Rinse the stopper and the sides of the flask using 50 ml distilled water.
10. Titrate the “TEST” against standardized sodium thiosulphate solution until a pale straw colour is
observed.
11. Add about 1ml starch indicator into the contents in the flask, a purple colour is observed.
12. Continue the titration until the color of the solution in the flask turns colourless.
13. The disappearance of the blue colour is recorded as the end point of the titration.
14. Similarly, the procedure is repeated for the flask labeled ‘Blank'.
15. Record the endpoint values of the BLANK .
16. Calculate the iodine number using the equation below:
Volume of Sodium thiosulphate used = [Blank- Test] mL
Equivalent Weight of Iodine = 127g
Normality of sodium thiosulphate ( Na2S203) = 0.1M
Observations: (Procedure)
1.
2.
3.
4.
5.
6.
7.
8.
10mL KI is to be added to
the test solution
9.
10.
After the pale yellow color is
obtained, starch is added to
the test solution and then
titrated till solution becomes
colorless from bluish black.
The end point of the titration
is noted.
11.
Similar procedure is followed with
blank only that fat is not added to the
blank
12.
Calculation is performed in the
following format.
Observations (Simulator):
BLANK-
COCONUT OIL-
SESAME OIL-
OLIVE OIL
MUSTARD OIL-
Name of
oil
Volume of
blank
Coconut
Sesame
Olive
Mustard
65.1
65.1
65.1
65.1
Formula used-
Volume of
test
solution
63.7
43.8
51.7
47.2
Na2S2O3
used
1.399
21.299
13.399
17.899
Normality of
Sodium
thiosuplate(N)
0.1
0.1
0.1
0.1
Equivalent
weight of
Iodine
127
127
127
127
Iodine
Value
6.35
133.35
82.55
107.94
Result:
The iodine value of coconut oil is 6.35
The iodine value of sesame oil is 133.35
The iodine value of olive oil is 82.55
The iodine value of mustard oil is 107.94
EXPERIMENT 5
Objective:
To characterize carbohydrates present in an unknown solution on the basis of various chemical
assays.
Theory:
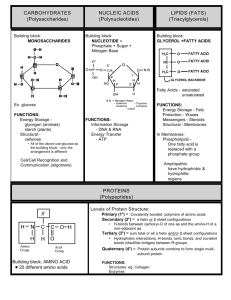
Carbohydrates are polyhydroxy aldehydes and ketones or substances that hydrolyze to yield
polyhydroxy aldehydes and ketones. Aldehydes (–CHO) and ketones ( = CO) constitute the major
groups in carbohydrates.
Fig:
Carbohydrates are mainly divided into monosaccharides, disaccharides and polysaccharides. The
commonly occurring monosaccharides includes glucose, fructose, galactose, ribose, etc. The two
monosaccharides combine together to form disaccharides which include sucrose, lactose and
maltose. Starch and cellulose fall into the category of polysaccharides, which consist of many
monosaccharide residues.
1.
Molisch’s Test:
This is a common test for all carbohydrates larger than tetroses. The test is on the basis that
pentoses and hexoses are dehydrated by conc. Sulphuric acid to form furfural or
hydroxymethylfurfural, respectively. These products condense with α-naphthol to form purple
condensation product.
2.
Fehling’s Test:
This forms the reduction test of carbohydrates. Fehling’s solution contains blue alkaline cupric
hydroxide solution, heated with reducing sugars gets reduced to yellow or red cuprous oxide and
is precipitated. Hence, formation of the yellow or brownish-red colored precipitate helps in the
detection of reducing sugars in the test solution.
3.
Benedict’s Test:
As in Fehling’s test, free aldehyde or keto group in the reducing sugars reduce cupric hydroxide
in alkaline medium to red colored cuprous oxide. Depending on the concentration of sugars, yellow
to green color is developed . All monosaccharides are reducing sugars as they all have a free
reactive carbonyl group. Some disaccharides, like maltose, have exposed carbonyl groups and are
also reducing sugars, but less reactive than monosaccharides
4.
Barfoed’s test :
Barfoed's test is used to detect the presence of monosaccharide (reducing) sugars in solution.
Barfoed's reagent, a mixture of ethanoic (acetic) acid and copper(II) acetate, is combined with the
test solution and boiled. A red copper(II) oxide precipitate is formed will indicates the presence of
reducing sugar. The reaction will be negative in the presence of disaccharide sugars because they
are weaker reducing agents. This test is specific for monosaccharides . Due to the weakly acidic
nature of Barfoed's reagent, it is reduced only by monosaccharides.
5.
Seliwanoff’s Test:
It is a color reaction specific for ketoses. When conce: HCl is added. ketoses undergo dehydration
to yield furfural derivatives more rapidly than aldoses. These derivatives form complexes with
resorcinol to yield deep red color. The test reagent causes the dehydration of ketohexoses to form
5-hydroxymethylfurfural. 5-hydroxymethylfurfural reacts with resorcinol present in the test
reagent to produce a red product within two minutes (reaction not shown). Aldohexoses reacts so
more slowly to form the same product.
6.
Bial’s Test:
Bial’s test is used to distinguish between pentoses and hexoses. They react with Bial’s reagent
and are converted to furfural. Orcinol and furfural condense in the presence of ferric ion to form a
colored product. Appearance of green colour or precipitate indicates the presence of pentoses and
formation of muddy brown precipitate shows the presence of hexoses.
7.
Iodine Test:
This test is used for the detection of starch in the solution. The blue-black colour is due to the
formation of starch-iodine complex. Starch contain polymer of α-amylose and amylopectin which
forms a complex with iodine to give the blue black colour.
8.
Osazone Test:
The ketoses and aldoses react with phenylhydrazine to produce a phenylhydrazone which further
reacts with another two molecules of phenylhydrazine to yield osazone. Needle-shaped yellow
osazone crystals are produced by glucose, fructose and mannose, whereas lactosazone produces
mushroom shaped crystals. Crystals of different shapes will be shown by different osazones.
Flower-shaped crystals are produced by maltose.
Materials Required:
1) Glassware
2) Test tubes
3) Test tube holder
4) Water bath
5) Spatula
6) Dropper
Reagents Required:
1) Molisch’s Reagent
2) Iodine solution
3)
4)
5)
6)
7)
8)
9)
10)
11)
12)
13)
Fehling’s reagent A
Fehling’s reagent B
Benedict’s qualitative reagent
Barfoed’s reagent
Seliwanoff ’s reagent
Bial’s reagent
Phenylhydrazine hydrochloride
Sodium acetate
Glacial acetic acid
Glucose, fructose
Microscope
Procedure:
No.
Test
Observation
Inference
Reaction
Molisch’s Test
1
2-3 drops of betanaphthol solution
are added to 2ml
A deep
of
the
test violet coloration is
Presence of
solution.
Very produced at the
carbohydrates.
gently add 1ml of junction of two
Conc.
layers.
H2SO4 along the
side of the test
tube..
This is due to the
formation of an
unstable
condensation
product of betanaphthol
with
furfural (produced
by the dehydration
of
the
carbohydrate).
Iodine test
4-5 drops of iodine
solution are added
Blue colour
2 to 1ml of the test
observed.
solution
and
contents are mixed
gently.
Fehling's test
3
About 2 ml of
sugar solution is
Iodine
forms
coloured
is Presence
of
adsorption
polysaccharide.
complexes
with
polysaccharides.
A red precipitate is Presence
formed
reducing sugar
This is due to the
of formation
of
cuprous oxide by
the reducing action
added to about 2
ml of Fehling’s
solution taken in a
test-tube. It is then
boiled for 10 min
of
the
sugar.
Benedict’s test
4
To 5 ml of
Benedict's
solution, add 1ml
of the test solution
If the saccharide is
and shake each
a reducing sugar it
Formation of a
tube. Place the
Presence of
will reduce Copper
green,
red,
or
tube in a boiling
reducing sugars [Cu] (11) ions to
yellow precipitate
water bath and
Cu(1) oxide, a red
heat
for
3
precipitate
minutes. Remove
the tubes from the
heat and allow
them to cool.
Barfoed’s test
5
To 2 ml of the
solution to be
tested added 2 ml
of
freshly
prepared
Barfoed's
reagent. Place test
tubes
into
a
boiling water bath
and heat for 3
minutes. Allow to
cool.
A deep blue colour
is formed with a red
ppt. settling down at
the bottom or sides
of the test tube.
Presence
of
reducing
sugars.
Appearance of a
red ppt as a thin
film at the bottom
of the test tube
within 3-5 min. is
indicative
of
reducing
monosaccharide. If the
ppt formation takes
more time, then it is
a
reducing
disaccharide.
If the saccharide is
a reducing sugar it
will reduce Cu (11)
ions to Cu(1) oxide
6
When reacted with
Seliwanoff reagent,
ketoses
react
within 2 minutes
forming
a
cherry
red
condensation
product
A
cherry
red
Presence
of
colored precipitate
ketoses
Seliwanoff test
within 5 minutes is
[Sucrose gives a
obtained.
To 3ml of of
positive ketohexose
Seliwanoff’s
test
]
reagent, add 1ml
of
the
test
solution. Boil in
Aldopentoses react
water bath for 2
slowly, forming the
A
faint
red
minutes.
Presence of aldoses coloured
colour
produced
condensation
product.
Bial's test
7
A
blue-green Presence
product
pentoses.
Add 3ml of Bial’s
reagent to 0.2ml of
the test solution. A muddy brown to Presence
Heat the solution
gray
product hexoses.
in a boiling water
bath for 2 minutes.
of
The
furfurals
formed produces
of condensation
products
with
specific colour.
Osazone Test
8
Two two ml of the
test solution, add
3ml of phenyl
hydrazine
hydrochloride
solution and mix.
Keep in a boiling
water bath for
30mts. Cool the
solution
and
observe
the
crystals
under
microscope.
Formation
beautiful
crystals of
Needle
crystals
Hedgehog
Sunflower
crystals
of
yellow
osazone Glucose/fructose
shaped
Reducing sugars
Presence of lactose
forms ozazone on
crystals Presence
of
treating
with
shaped maltose
phenylhydrazine
Observation:
1. MOLISCH TEST
2. BENEDICT’S TEST
3. FEHLING’S TEST
4. BARFOED’S TEST
5. SELIWANOFF’S TEST
6. BIAL’S TEST
7. IODINE TEST
8. OSAZONE TEST
Result:
Conclusion: The given sugar maybe ribose
.
EXPERIMENT 6
Objective:
To perform the isoelectric precipitation of casein present in milk.
Theory:
Milk is a mixture of many types of proteins, most of them present in very small amounts. Milk
proteins are classified into three main groups of proteins on the basis of their widely different
behaviors and forms of existence. They are caseins (80%), whey proteins and minor proteins.
Casein is a heterogeneous mixture of phosphorous containing proteins in milk. Casein is present
in milk as calcium salt and calcium caseinate. It is a mixture of alpha, beta and kappa caseins to
form a cluster called micelle. These micelles are responsible for the white opaque appearance of
milk.
Casein, like proteins, are made up of many hundreds of individual amino acids. Each may
have a positive or a negative charge, depending on the pH of the [milk] system. At some pH
value, all the positive charges and all the negative charges on the [casein] protein will be in
balance, so that the net charge on the protein will be zero.
That pH value is known as the isoelectric point (IEP) of the protein and is generally the pH at
which the protein is least soluble. For casein, the IEP is approximately 4.6 and it is the pH value
at which acid casein is precipitated. In milk, which has a pH of about 6.6, the casein micelles
have a net negative charge and are quite stable. During the addition of acid to milk, the negative
charges on the outer surface of the micelle are neutralized (the phosphate groups are protonated),
and the neutral protein precipitates.
The same principle applies when milk is fermented to curd. The lactic acid bacillus produces
lactic acid as the major metabolic end-product of carbohydrate [lactose in milk] fermentation.
The lactic acid production lowers the pH of milk to the IEP of casein. At this pH, casein
precipitates.
Materials required:
1) Raw milk - 100ml
2) 0.2N HCl - 50ml
3) Diethyl ether - 50ml
4) 50% Ethanol - 50ml
5) Whatman No 1 filter paper strip (Size 25×50mm) - 2 no.
Procedure:
1. Measure 100ml of milk in a measuring cylinder and transfer 25ml of milk to four Oakridge
centrifuge tubes each.
2. Centrifuge the milk in a centrifuge at 4000rpm at room temperature (25- 30o C) for 20 minutes.
This is done to remove the fats and lipids from the mixture.
3. After centrifugation, carefully remove the fats and lipids from the surface of the milk with a
spatula.
4. Then transfer the milk from all the tubes into a beaker and add equal volume of distilled water and
stir well. Now check the pH.
5. Start adding 0.2N HCl drop by drop into the milk mixture and stir well.
6. Note the PH at which precipitation (white curdy substances) appears. The pH should be 4.6.
7. Take the curdy precipitate and allow it to sediment.
8. Now decant the supernatant using a filter paper and funnel and wash the precipitate with distilled
water to remove the salts, then wash with diethyl ether and ethanol.
9. Dry the precipitate and take the weight of the casein and record it.
Observations (Animation video):
1
2
1
3
2
2
2
1
4
3
3
2
2
2
1
5
3
3
2
2
2
1
6
3
3
2
2
2
1
7
3
3
2
2
2
1
Simulator:
Result:
Casein gets precipitated at pH 4.6 where it has the least solubility. Thus 4.6 is the isolectric pH
of casein.
EXPERIMENT 7
Aim: To extract DNA from leaves/ fruit at home.
Theory: Plant materials are among the most difficult for high quality DNA extractions. The
key is to properly prepare the tissues for extraction. In most cases this involves the use of
liquid nitrogen flash freezing followed by grinding the frozen tissue with a mortar and pestle.
Liquid nitrogen is difficult to handle and it is dangerous in an open laboratory environment
such as a classroom. For this reason, we have modified a very simple plant DNA extraction
protocol to use fresh tissue.
Materials Required:
o
o
o
o
o
o
o
o
o
o
Fruit – Kiwi, Strawberries, and Banana, leafy vegetables all work well
5 g washing up liquid
2 g salt
100 ml tap water
100 ml of ice-cold alcohol (isopropyl alcohol can usually be found at the pharmacists);
put in a freezer for at least 30 mins before starting the experiment
Access to hot water – about 60 °C
Sieve or coffee filter paper
Two glass beakers (or old jam-jars)
Several bowls of different sizes, including a large bowl for making a water bath
Freshly ground papaya juice/ pineapple juice
Procedure:
o Mash up the fruit/leaves of your choice in a bowl. This experiment uses spinach leaves.
o In a separate bowl, mix the washing up liquid, salt and tap water. Stir gently trying to
avoid making too many bubbles in the mixture. This is your extraction buffer.
o Add the fruit to the extraction buffer and mix again. Mash your sample as much as you
can, but again, try to avoid making too many bubbles.
o Make a water bath with a temperature of about 60 °C. (A large washing up bowl works
well for this.) Leave the extraction mixture to incubate for 15 minutes.
o After 15 minutes, filter your mixture through a fine sieve or coffee filter. This will
remove all the solid material that you don’t want. You should be left with a clear(ish)
liquid.
o Take the ice-cold alcohol and very slowly, drop by drop, pour it down the inside of the
container with your fruit mixture. What you want to do is produce a layer of the alcohol
floating on top of the mixture.
o At the interface between the alcohol and the mixture, you should see a white cloud-like
substance forming. Use a hook (a bent paperclip would work) to slowly draw the DNA
up and out of the solution.
Observations: Material used- Spinach leaves
Result: DNA was successfully extracted from spinach leaves
EXPERIMENT 8
Objective:
To retrieve more information about the drug molecules, drug targets, enzymes and pathways
related to drugs.
Theory:
Drug is a chemical molecule that can interact and bind to a target and control the function
biological receptors to control the disease. Proteins are the important biological receptors which
interact with the other biological molecules to maintain various cellular functions. Drugs have a
specific activity that goes and binds to the target and can activate/inhibit the reactions in a proper
manner. To find more about the drug molecules, their target molecules and the drug enzymatic
reactions and the drug involvement in a specific pathway can easily found using different databases
like Drugbank.
Drugbank is a freely accessible, web enabled, fully query based database. It contains a data like
structure and activity of a drugs and structure/function/sequence of a protein molecule. Drugbank
is unique bioinformatics and cheminformatics database, having the details of drug molecules and
their target molecules. It gives well developed concepts search and also comparisons from
medicinal chemistry. Database will be updated by semi-annually.
Drugbank database contains 6711 drug entries. It includes 1447 FDA-approved small drugs, 131
FDA-approved protein/peptide drugs, 85 nutraceuticals and 5080 experimental drugs.
Additionally, 4227 non-redundant protein sequences are linked to these drug entries.
Each DrugCard entry contains more than 150 data fields with half of the information being devoted
to drug/chemical data and the other half devoted to drug target or protein data.
Searching through drug bank can also be supported with the help of logical operators. Keeping the
query word in quotes can help to search exact match, in a search by giving “acetic acid” will allow
search for only “acetic acid” and not its similar matches. Another type of search can be done by
using wild cards is by giving * with the query word, the result would be hits which starts with the
query word.
In Drugbank browse option one can get the drug molecules by using “Drug browse”. It allows the
user to get all the drug molecules which has been stored in the database by alphabetically with an
information like Drugbank ID, CAS number, Molecular weight, Structure, Categeories,
Therapeutic indication. “Pharma browse “ allows the user to find the drug molecules by categeory
wise. Geno browse gives the chemical molecules with Drug name, with which enzyme it is going
to bind, SNPs, all the adverse reactions and references belongs to the Drug molecule. “pathway
Browse” allows one to search the pathway which is going to affect by the drug molecule. “Class
Browse” allows one to search the drug classes by names. “Association browse” allows user to find
the name of the drug, Genename , which species it belongs and its specific function.
Drug bank gives the details about the drugs with information like:
Table 1: Information regarding Drugs.
Generic Name
name
DrugBank ID
drugbank_id
AHFS Code
ahfs_code
ATC Code
atc_code
Absorption
absorption
Affected Organisms organism
Chemical Formula
formula
Chemical
Name
iupac
IUPAC
Clearance
clearance
DPD ID (DIN)
dpd_id
Description
description
Dosage Form
dosage_form
Dosage Route
dosage_route
Drug Group
group
Drug Type
type
Drug-Food
Interaction
food_interaction
Genbank ID
genbank_id
Guide
to
guide_to_pharmacology_id
Pharmacology ID
H2O
Solubility
experimental_water_solubility
(experimental
HET ID
het_id
Brands
brand
CAS Number
cas_number
Caco2 Permeability
experimental_caco2_permeability
(experimental)
Category
category
ChEBI ID
chebi_id
Half Life
half_life
ID
id
IUPHAR ID
iuphar_id
InChI Identifier
inchi
InChI Key
inchikey
Indication
indication
KEGG Compound
kegg_compound_id
ID
KEGG Drug ID
kegg_drug_id
LogP
(experimental)
experimental_logp
LogP (predicted)
predicted_logp
LogS
(experimental)
experimental_logs
LogS (predicted)
predicted_logs
Mechanism
Action
of
mechanism_of_action
Melting Point (°C)
melting_point
Metabolism
metabolism
Mixture
Names
Brand
Mixture
Ingredients
Molecular
(average)
mixture_name
mixture_ingredient
Weight
average_mass
PDB ID
pdb_id
PDRhealth Link
pdrhealth_link
Pathway
ID
pathway_smpdb_id
SMPDB
Pathways
pathway_name
PharmGKB ID
pharmgkb_id
Pharmacodynamics pharmacodynamics
Protein Binding
protein_binding
PubChem
Compound ID
pubchem_compound_id
Route
Elimination
of
route_of_elimination
RxList Link
rxlist_link
SMILES
smiles
Secondary
secondary_id
Accession Number
State
state
Synonyms
synonym
Target ID
target_id
Toxicity
toxicity
pKa/Isoelectric
Point
pka_isoelectric_point
UniProt Name
uniprot_name
UniProt ID
uniprot_id
Wikipedia Link
wikipedia_link
Observations:
1. Azithromycin: link- https://go.drugbank.com/drugs/DB00207
Generic Name
Azithromycin
DrugBank ID
DB00207
52:04.04 — Antibacterials
AHFS Code
08:12.12.92 — Other Macrolides
ATC Codes
J01RA07 — Azithromycin, fluconazole and secnidazole
J01RA — Combinations of antibacterials
J01R — COMBINATIONS OF ANTIBACTERIALS
J01 — ANTIBACTERIALS FOR SYSTEMIC USE
J — ANTIINFECTIVES FOR SYSTEMIC USE
S01AA26 — Azithromycin
S01AA — Antibiotics
ATC Code
S01A — ANTIINFECTIVES
S01 — OPHTHALMOLOGICALS
S — SENSORY ORGANS
J01FA10 — Azithromycin
J01FA — Macrolides
J01F — MACROLIDES, LINCOSAMIDES AND STREPTOGRAMINS
J01 — ANTIBACTERIALS FOR SYSTEMIC USE
J — ANTIINFECTIVES FOR SYSTEMIC USE
Absorption
Affected Organisms
Bioavailability of azithromycin is 37% following oral administration. Absorption is not affected by foo
Macrolide absorption in the intestines is believed to be mediated by P-glycoprotein (ABCB1) efflux
transporters, which are known to be encoded by the ABCB1 gene 4.
Humans and other mammals
Chemical Formula
Chemical IUPAC
Name
Clearance
C38H72N2O12
(2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-11-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-
methyloxan-2-yl]oxy}-2-ethyl-3,4,10-trihydroxy-13-{[(2R,4R,5S,6S)-5-hydroxy-4-methoxy-4,6dimethyloxan-2-yl]oxy}-3,5,6,8,10,12,14-heptamethyl-1-oxa-6-azacyclopentadecan-15-one
Mean apparent plasma cl=630 mL/min (following single 500 mg oral and i.v. dose)
DRG-0104
DPD ID (DIN)
Azithromycin is a broad-spectrum macrolide antibiotic with a long half-life and a high degree of tissue
penetration 3. It was initially approved by the FDA in 1991.
It is primarily used for the treatment of respiratory, enteric and genitourinary infections and may be use
instead of other macrolides for some sexually transmitted and enteric infections. It is structurally related
to erythromycin.
Description
Azithromycin [9-deoxo-9a-aza-9a-methyl-9a-homoerythromycin] is a part of the azalide subclass of
macrolides, and contains a 15-membered ring, with a methyl-substituted nitrogen instead of a carbonyl
group at the 9a position on the aglycone ring, which allows for the prevention of its metabolism. This
differentiates azithromycin from other types of macrolides.
In March 2020, a small study was funded by the French government to investigate the treatment of
COVID-19 with a combination of azithromycin and the anti-malaria drug hydroxychloroquine. The
results were positive, all patients taking the combination were virologically cured within 6 days of
treatment, however, larger studies are required.9
Dosage Form
Drug Group
Anti-Bacterial Agents
Anti-Infective Agents
Antibacterials for Systemic Use
Antiinfectives for Systemic Use
Drug Type
Small Molecule
Drug-Food
Interaction
Take on an empty stomach. Take at least 1 hour before or 2 hours after meals.
Genbank ID
ABCB1
Guide to
Pharmacology ID
MQTOSJVFKKJCRP-BICOPXKESA-N
H2O Solubility
(experimental
0.514 mg/mL
CP 62993
HET ID
Brands
Act Azithromycin
UniProt Name
Protein-arginine deiminase type-4
UniProt ID
http://www.uniprot.org/uniprot/Q9UM07
Wikipedia Link
https://en.wikipedia.org/wiki/Azithromycin
2. Folic acid:
3. Mebutizide:
4. Ibuprofen:
EXPERIMENT 9
Aim
To prepare a temporary mount of onion root tip to study mitosis.
Theory
All organisms are made of cells. For an organism to grow, mature and maintain tissue, new cells
must be made. All cells are produced by division of pre-existing cells. Continuity of life depends
on cell division. There are two main methods of cell division: mitosis and meiosis. In this tutorial
we will learn about mitosis.
What is Mitosis?
Mitosis is very important to life because it provides new cells for growth and replaces dead cells.
Mitosis is the process in which a eukaryotic cell nucleus splits in two, followed by division of the
parent cell into two daughter cells. Each cell division consists of two events: cytokinesis and
karyokinesis. Karyokinesis is the process of division of the nucleus and cytokinesis is the process
of division of cytoplasm.
Events during Mitosis
1. Prophase:
1. Mitosis begins at prophase with the thickening and coiling of the chromosomes.
2. The nuclear membrane and nucleolus shrinks and disappears.
3. The end of prophase is marked by the beginning of the organization of a group of
fibres to form a spindle.
2. Metaphase
1. The chromosome become thick and two chromatids of each chromosome become
clear.
2. Each chromosome attaches to spindle fibres at its centromere.
3. The chromosomes are arranged at the midline of the cell.
3. Anaphase
1. In anaphase each chromatid pair separates from the centromere and move towards
the opposite ends of the cell by the spindle fibres.
2. The cell membrane begins to pinch at the centre.
4. Telophase
1. Chromatids arrive at opposite poles of cell.
2. The spindle disappears and the daughter chromosome uncoils to form chromatin
fibres.
3. The nuclear membranes and nucleolus re-form and two daughter nuclei appear at
opposite poles.
4. Cytokinesis or the partitioning of the cell may also begin during this stage.
The stage, or phase, after the completion of mitosis is called interphase. It is the non dividing
phase of the cell cycle between two successive cell divisions. Mitosis is only one part of the cell
cycle. Most of the life of a cell is spent in interphase. Interphase consist of three stages call G1, S
and G2.
Mitosis in Onion Root Tip
The meristamatic cells located in the root tips provide the most suitable material for the study of
mitosis. The chromosome of monocotyledonous plants is large and more visible, therefore, onion
root tips are used to study mitosis. Based on the kind of cells and species of organism, the time
taken for mitosis may vary. Mitosis is influenced by factors like temperature and time
Observations:
40X (Low Power)
100X (High Power)
Result: Mitosis in onion root tip cells was successfully studied at low (40X) and high
(100X) magnification.
EXPERIMENT 10
Aim: To find the acid value of given sample of coconut oil
Theory: Rancidity may occur in CMM samples upon storage especially when CMM contains high
content of fatty acid or fatty oils. The decomposed components such as free fatty acids, peroxides,
low molecular weight aldehydes and low molecular weight of ketones are produced. This would
result in distinctive smell and affect the quality of the CMM samples. In view of this, acid value
which is defined as the number of mg of potassium hydroxide required to neutralize the free acid
in 1g of fat, fatty oil or other related substances is determined to assess the rancidity of the CMM
samples. Sodium hydroxide may also be used.
Reagents: Coconut oil, Phenolphthalein indicator, Sodium hydroxide/ potassium hydroxide
titrant, Ethanol-ether solution
Method and procedure:
o Take sample of coconut oil (2.5 g) in a conical flask (250mL)
o Weigh accurately 0.56g of KOH and place it in a 1000-mL volumetric flask and make 100
mL solution with distilled water.
o Prepare a 50mL mixture of ethanol and diethyl ether (1:1, v/v). Dissolve the coconut oil in
the mixture. Shake it well. If necessary, reflux the mixture gently until the substance is
completely dissolved.
o Standardize the NaOH/ KOH solution prepared with potassium hydrogen phthalate.
o The solution of KOH prepared was 0.1M and was successfully standardized.
o Titrate the ethanol mixture with KOH till using phenolphthalein indicator. This is the blank
o Titrate the sample with potassium hydroxide titrant until pink coloration can be observed
which persists for 30 s. This is the reading for the sample called test.
(KOH)
Observations:
Weight of oil: 2.5 g
Average blank reading: 0.9 mL
Average test reading: 1.0 mL
Volume of KOH used by coconut oil sample: (1-0.9) = 0.1 mL
Acid Value: (0.1 mL × 5.6)/ 2.5 = 0.2244 mg
Result: Acid value for the given sample of coconut oil was found to be 0.2244 mg KOH.