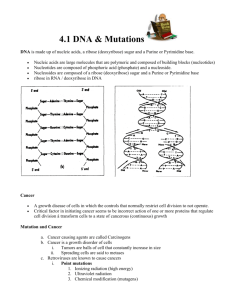
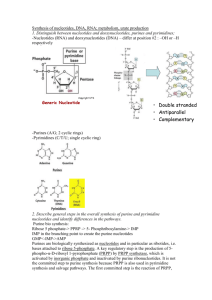
BMS204 Nucleotide Metabolism Dr. Reem Arafa Professor of Biomedical Sciences Central Dogma of Biology Nucleic Acids Nucleic Acids Are Essential For Information Transfer in Cells Information encoded in a DNA molecule is transcribed via synthesis of an RNA molecule The sequence of the RNA molecule is "read" and is translated into the sequence of amino acids in a protein. Compounds containing C, N, O, and high amount of P. Was an acid compound found in nuclei therefore named nucleic acid 1944 Oswald, Avery, MacLeod and McCarty demonstrated that DNA is the molecular carrier of genetic information. 1953 Watson and Crick proposed the double helix model for the structure of DNA Nucleic acids as long polymers of nucleotides. Nucleotides and Nucleic Acids The amino acids sequence in a protein and nucleotide sequence in RNA is specified by a nucleotide sequence in the cell’s DNA Gene: segment of a DNA molecule that contains the information required for the synthesis of a functional biological product whether a protein or RNA The cell contains thousands of genes and DNA molecules Nucleic Acids are polymers of nucleotides. Deoxyribonucleic acid - DNA - Storage of genetic information. Ribonucleic acid - RNA - carriers of genetic information and catalysis. Nucleotide Structure: Nitrogenous base + ribose + phosphate Nucleotides structure The nucleotide has three characteristic components Nitrogenous base Pentose sugar Phosphate Nitrogenous Bases Nitrogenous base: derivatives of Purines and pyrimidines DNA and RNA contain the same purine bases and the pyrimidine base Cytosine but Thymine is found only in DNA and Uracil is found only in RNA (A) (C) (G) (U) (T) Pentoses of Nucleotides • • • • D-ribose (in RNA) 2-deoxy-D-ribose (in DNA) The difference is 2'-OH vs 2‘-H This difference affects secondary structure and stability Bases are attached by β-N-glycosidic linkages to carbon 1 of pentose sugar – (Nucleoside) • Base is linked via a β-N-glycosidic bond • The carbon of the glycosidic bond is anomeric • Named by adding -idine to the root name of a pyrimidine or -osine to the root name of a purine • Sugars make nucleosides more water-soluble than free bases Nucleosides (1) (9) Nucleotides • Phosphate ester of nucleosides Nucleotides The plane of the base is oriented perpendicular to the plane of the pentose group RNA contains Ribose while DNA contains 2'deoxy-D-Ribose Ribonucleotides Adenosine 5'-monophosphate, Adenylate, AMP Guanosine 5'-monophosphate, Guanylate, GMP Cytidine 5'-monophosphate, Cytidylate, CMP Uridine 5'-monophosphate, Uridylate, UMP Deoxyribonucleotides Deoxythymidine 5‘-monophosphate, Deoxythymidine, dTMP …….. NMP=== Nucleoside mono phosphate. Numbering of Ribose sugar is given 1’, 2’, …, 5’ Anti- conformation predominates in nucleic acid polymers • Conformation can be syn or anti Unusual nucleotides Modified nucleotides are found in some viral DNA and in Transfer RNA. These modifications include methylation, hydroxymethylation, glycosylation, acetylation. Other Functions of Nucleotides • Nucleoside 5'-triphosphates are carriers of energy • Bases serve as recognition units • Cyclic nucleotides are signal molecules and regulators of cellular metabolism and reproduction e.g. cyclic AMP • Structural component of some coenzymes, e.g. CoA, FAD, NADH, NADPH • ATP is central to energy metabolism • GTP drives protein synthesis • CTP drives lipid synthesis • UTP drives carbohydrate metabolism De novo synthesis of purine nucleotides The atoms of purine ring are contributed by a number of compounds including amino acids (aspartic acid, glycine and glutamine), CO2, and tetrahydrofolate. These compounds donates N and C to constructed Ribose 5-phosphate. 6 5 1 2 4 3 7 8 9 N10Formyl tetrahydrofolate The order in which ring atoms are added is: 9 Glut 4 5 7 Glycine 8 For 3 6 Glut 1 CO2 6 5 1 N 2 2 Asp N 7 8 N 3 4 N 9 Ribose-P For De novo synthesis of purine nucleotides * Synthesis of 5-phosphoribosyl-1-pyrophosphate (PRPP) - Ribose 5-phosphate is synthesized from HMP (Hexose monophosphate pathway) - Ribonucleotides are first synthesized then reduced to deoxyribonucleotides Inhibitors Purines, nucleosides Activator Pi Ribose-5-phosphate Ribose phosphate pyrophosphokinase ATP AMP Synthesis of 5-phospho-β-D-ribosylamine •The amide group of the glutamine replaces pyrophosphate group attached to PRPP, this reaction is mediated by Glutamine:phosphoribosyl pyrophosphate amidotransferase. •This enzyme is inhibited by end product of this pathway purine 5’-nucleotides AMP, GMP and IMP. • This reaction is the committed step in purine nucleotide synthesis De novo synthesis of purine nucleotides Synthesis Inosine monophosphate (IMP) • IMP is the parent purine nucleotide • Synthesis of IMP requires 4 ATP molecules Inhibitors of Purine synthesis • Specific inhibitors that inhibits the growth of rapidly growing microorganisms e.g. Sulfonamides • Structural analogues for folic acid (methotroxate) * Conversion of IMP into AMP and GMP - this reaction is energy-requiring pathway Adenylosuccinate synthetase Feed back inhibition IMP dehydrogenase Feed back inhibition Conversion of IMP into AMP and GMP Conversion of Nucleoside momophosphate (NMP) to nucleoside diphosphate (NDP) and triphosphate (NTP) • NDP and NTP are synthesized from the corresponding NMP by Nucleoside monophosphate kinases • These kinases don't discriminate between ribose or deoxyribose in the substrate. • ATP is the source of the transferred phosphate examples AMP + ATP 2 ADP Adenylate kinase (highly active in the liver) GMP + ATP GDP + ADP Guanylate kinase • Nucleoside diphosphates and triphosphates are interconverted by nucleoside diphosphate kinase GDP + ATP GTP + ADP CDP + ATP CTP + ADP De novo synthesis of purine nucleotides Salvage Pathway for Purines Salvage Pathway: Purines that results from the normal turnover of cellular nucleic acids or obtained from the diet and not degraded can be reconverted into nucleoside triphosphates and used by the body. Two enzymes are involved: Adenine phosphoribosyl transferase (APRT) Hypoxanthine-guanine phosphoribosyl transferase (HGPRT) Both enzymes utilize PRPP as the source of ribose5-phosphate group Degradation of Purine nucleotides Degradation of Purine nucleotides Purines are sequentially degraded into uric acids (in humans) Several steps are involved in this catabolic pathway Degradation of Dietary nucleic acids in the small intestine • Ribonucleases and deoxyriboncleases secreted in the pancreatic juice can hydrolyze RNA and DNA into oligonucleotides. • Oligonucleotides are further hydrolyzed by pancreatic phophodiesterases producing a mixture of 3’-and 5’ mononucleotides • A family of nucleotidases remove the phosphate group releasing nucleosides that get absorbed from GIT • Dietary nucleotides are not used to large extent in cells because they are converted into uric acid in the small intestine and also used by the normal flora Degradation of Purine nucleotides Pyrimidine nucleotides synthesis Sources of carbon atoms in pyrimidine rings - Purine ring is synthesized on an existing ribose 5-phosphate - Pyrimidine ring is synthesized then attached to ribose 5-phosphate donated by PRPP - the sources of carbon atoms in pyrimidine rings are Glutamine, CO2, and aspartic acid Aspartic Acid CO2 Pyrimidine nucleotides synthesis - Synthesis of carbamoyl phosphate - The committed step of this pathway in mammalian cell is the synthesis of carbamoyl phosphate from Glutamine and CO2 2 ATP + CO2 + Glutamine Carbamoyl phosphate + 2ADP + Glutamate - This reaction is mediated by Carbamoyl Phosphate Synthetase II (CPS II) - CPS II is inhibited by UTP and activated by ATP and PRPP - Carbamoyl phosphate is the precursor of Urea; the pyrimidine synthesis occurs in the cytosol while the urea production occurs in the mitochondria by Carbamoyl Phosphate Synthetase I (CPS I) - CPS I uses ammonia as source of nitrogen - CPS II uses the amide group of glutamine - Glutamine is required in the synthesis of both Purines and Pyrimidine Synthesis of carbamoyl phosphate De Novo Pyrimidine Synthesis De Novo Pyrimidine Synthesis Degradation of pyrimidines - Purines are not cleaved in human cell - Pyrimidines rings can be opened and degraded to highly soluble structures, such as -alanine and -aminoisobutyrate that can serve as precursors of acetyl CoA and succinyl CoA - Pyrimidine can be salvaged and converted into nucleotides by the enzyme Pyrimidine phosphoribosyltransferase and it utilizes the PRPP Conversion of Ribonucleotides to Deoxyribonucleotides 2’-deoxyribonucleotides are synthesized from ribonucleoside diphosphatase Ribonucleotide reductase is multi-subunit enzyme (2B1 and 2B2 subunits) that catalyzes the reduction of NDP (ADP, GDP, CDP, UDP) into dNDP (dADP, dGDP, dCDP, dUDP) The immediate donors of hydrogen atoms needed for the reduction are two–SH groups of the enzyme itself The reduced form of the enzyme should be regenerated The reducing agent is a peptide coenzyme of ribonucleotide reductase called Thioredoxin The thioredoxin contain two cysteine residues that can be oxidized to reduce the ribonucleotide reductase enzyme. The oxidized thioredoxin is reduced back by NADPH, this reaction is mediated by Thioredoxine reductase Regulation of Deoxyribonucleotides synthesis The regulation of this enzyme is complex Not only the activity is regulated but also substrate specificity The binding of dATP to an allosteric site called activity site inhibits the enzyme while the binding of ATP to this site activate the enzyme. The binding of NTP to an allosteric site called substrate specificity will increase the conversion of different NTP to their corresponding dNTP according to the need of the cell When ATP or dATP are bound to this site reduction of UDP, and CDP is favored When dTTP,or dGTP is bound the reduction of GDP, ADP is stimulated Synthesis of deoxythymidine monophosphate from dUMP - dUMP is converted into dTMP by thymidylate synthetase which utilizes N5,N10 –methylene tetrahydrofolate as the source of methyl group and 2 hydrogen oxidation into dihydrofolate. - inhibitors of thymidylate synthetase (5-flouro uracil) act as anti-tumor - DHF can be reduced into THF by DHF reductase, which can be inhibited by Methotrexate inhibits purine synthesis and decrease the supply of THF so prevents methylation of dUMP to dTMP The End