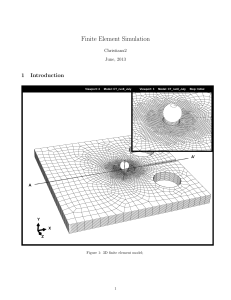
Dynamic Causal Modelling (DCM):
Theory
Demis Hassabis & Hanneke den Ouden
Thanks to
Klaas Enno Stephan
Functional Imaging Lab
Wellcome Dept. of Imaging Neuroscience
Institute of Neurology
University College London
Overview
• Classical approaches to functional & effective
connectivity
• Generic concepts of system analysis
• DCM for fMRI:
– Neural dynamics and hemodynamics
– Bayesian parameter estimation
• Interpretation of parameters
– Statistical inference
– Bayesian model selection
System analyses in functional
neuroimaging
Functional specialisation
Functional integration
Analyses of regionally specific effects:
which areas constitute a neuronal
system?
Analyses of inter-regional effects:
what are the interactions between the
elements of a given neuronal system?
Functional connectivity
Effective connectivity
= the temporal correlation between
spatially remote neurophysiological
events
MECHANISM-FREE
= the influence that the elements of a
neuronal system exert over another
MECHANISTIC
Models of effective connectivity
• Structural Equation Modelling (SEM)
• Psycho-physiological interactions (PPI)
• Multivariate autoregressive models (MAR)
& Granger causality techniques
• Kalman filtering
• Volterra series
• Dynamic Causal Modelling (DCM)
Friston et al., NeuroImage 2003
Overview
• Classical approaches to functional & effective
connectivity
• Generic concepts of system analysis
• DCM for fMRI:
– Neural dynamics and hemodynamics
– Bayesian parameter estimation
• Interpretation of parameters
– Statistical inference
– Bayesian model selection
Models of effective connectivity = system models.
But what precisely is a system?
• System =
set of elements which interact in a
spatially and temporally specific fashion.
• System dynamics =
change of state vector in time
• Causal effects in the system:
– interactions between elements
– external inputs u
• System parameters :
specify the nature of the interactions
• general state equation for nonautonomous systems
z1 (t ) overall
state
z (t ) = system
represented
by state variables
z n (t )
z1 f1 (zz11...z n , u, 1 )
of
dz
= change
state
vector
= z = in time
dt
zn f n(zz1...z n , u, n )
n
z = F ( z, u, )
Example:
linear
dynamic
system
FG
z3 left
z1
LG
left
RVF
u2
state
changes
z = Az + Cu
= { A, C}
FG
right
LG
right
z4
LG = lingual gyrus
FG = fusiform gyrus
z2
Visual input in the
- left (LVF)
- right (RVF)
visual field.
LVF
u1
effective
connectivity
system
state
input
external
parameters inputs
z1 a11 a12 a13 0 z1 0 c12
z c
z a a
u
0
a
0
1
24 2
21
2 = 21 22
+
z3 a31 0 a33 a34 z3 0 0 u2
z4 0 a42 a43 a44 z 4 0 0
Extension:
bilinear
dynamic
system
z3
FG
left
FG
right
z4
m
z = ( A + u j B j ) z + Cu
z1
RVF
u2
LG
left
LG
right
CONTEXT
u3
0 b123
z1 a11 a12 a13 0
z a a
0
a
0 0
24
2 = 21 22
+ u3
0 0
z3 a31 0 a33 a34
0 0
z4 0 a42 a43 a44
j =1
z2
LVF
u1
0 0 z1 0 c12
0 0 z2 c21 0
+
3
0 b34 z3 0 0
0 0 z4 0 0
0
u1
0
u2
0
u3
0
Bilinear state equation in DCM
state
changes
intrinsic
connectivity
modulation of system
connectivity
state
direct
inputs
m external
inputs
j
j
z1 a11 a1n m b11 b1n z1 c11 c1m u1
= + u +
j
j
=
1
j
j
bn1 bnn zn cn1 cnm um
zn an1 ann
m
z = ( A + u j B ) z + Cu
j
j =1
Overview
• Classical approaches to functional & effective
connectivity
• Generic concepts of system analysis
• DCM for fMRI:
– Neural dynamics and hemodynamics
– Bayesian parameter estimation
• Interpretation of parameters
– Statistical inference
– Bayesian model selection
DCM for fMRI: the basic idea
• Using a bilinear state equation, a cognitive system is
modelled at its underlying neuronal level (which is not
directly accessible for fMRI).
• The modelled neuronal dynamics (z) is transformed into
area-specific BOLD signals (y) by a hemodynamic
forward model (λ).
The aim of DCM is to estimate parameters at the
neuronal level such that the modelled BOLD signals
are maximally similar to the experimentally measured
BOLD signals.
z
λ
y
Conceptual
overview
Neural state equation
z = F ( z, u, n )
The bilinear model
z = ( A + u j B j ) z + Cu
F z
=
z z
2F
z
j
B =
=
zu j u j z
A=
effective connectivity
modulation of
connectivity
Input
u(t)
c1
C=
direct inputs
b23
a12
activity
z2(t)
activity
z1(t)
integration
neuronal
states
activity
z3(t)
z
λ
y
y
F z
=
u u
hemodynamic
model
y
BOLD
y
Friston et al. 2003,
NeuroImage
Example:
generated neural
data
stimuli
u1
context
u2
+
u1
u1
u2
u2
Z1
z1
Z2
-
Z1
+
z2
+
z = Az + u2 B 2 z + Cu1
Z2
-
2
z
a
12
1
b
11
=
z
+
u
2
z a 21
0
2
0 c1 0 u1
2 z +
u
0
0
b22 2
The hemodynamic “Balloon” model
• 5 hemodynamic
parameters:
activity
z(t )
= { , , , , }
h
vasodilatory signal
s = z − s − γ( f − 1)
s
f
flow induction
important for model fitting,
but of no interest for
statistical inference
• Empirically determined
a priori distributions.
• Computed separately for
each area (like the neural
parameters).
f = s
f
changes in volume
τv = f − v
1 /α
v
changes in dHb
τq = f E ( f, ) q − v1 /α q/v
q
v
BOLD signal
y (t ) = (v, q )
Example: modelled BOLD signal
Underlying model
left LG
(modulatory inputs not shown)
FG
left
FG
right
LG
left
LG
right
RVF
LG = lingual gyrus
FG = fusiform gyrus
right LG
LVF
Visual input in the
- left (LVF)
- right (RVF)
visual field.
blue:
red:
observed BOLD signal
modelled BOLD signal (DCM)
Overview
• Classical approaches to functional & effective
connectivity
• Generic concepts of system analysis
• DCM for fMRI:
– Neural dynamics and hemodynamics
– Bayesian parameter estimation
• Interpretation of parameters
– Statistical inference
– Bayesian model selection
Bayesian rule in DCM
Bayes Theorem
p( | y ) p( y | ) p( )
posterior
likelihood
∙ prior
• Likelihood derived from error
and confounds (eg. drift)
• Priors – empirical
(haemodynamic parameters)
and non-empirical (eg.
shrinkage priors, temporal
scaling)
• Posterior probability for each
effect calculated and
probability that it exceeds a set
threshold expressed as a
percentage
stimulus function u
Parameter estimation in DCM
z = ( A + u j B j ) z + Cu
• Combining the neural and
hemodynamic states
gives the complete
forward model.
• An observation model
includes measurement
error e and confounds X
(e.g. drift).
activity - dependent vasodilatory signal
s = z − s − γ( f − 1)
s
s
f
flow - induction (rCBF)
hidden states
x = {z , s, f , v, q}
h = { , , , , }
f
n = { A, B1...B m , C}
= { h , n }
x = F ( x, u, )
changes in volume v
τv = f − v1/α
v
ηθ|y
parameters
f = s
state equation
• Bayesian parameter
estimation: minimise
difference between data
and model
• Result:
Gaussian a posteriori
parameter distributions,
characterised by
mean ηθ|y and
covariance Cθ|y.
neural state
equation
changes in dHb
τq = f E ( f, ) q − v1/α q/v
q
y = (x )
y = h(u, ) + X + e
modelled
BOLD response
observation model
Overview
• Classical approaches to functional & effective
connectivity
• Generic concepts of system analysis
• DCM for fMRI:
– Neural dynamics and hemodynamics
– Bayesian parameter estimation
• Interpretation of parameters
– Statistical inference
– Bayesian model selection
DCM parameters:
interpretation & inference
- DCM gives gaussian
posterior densities of
parameters (intrinsic
connectivity, effective
connectivity and inputs)
–How can we make inference
about effects represented by
these parameters
Hypothesis: modulation by context > 0
z3 left
FG
FG
right
z4
LG
left
LG
right
z2
z1
–At a single subject level?
–At a group level?
– How do we select between
different models?
RVF
u2
CONTEXT
u3
LVF
u1
Bayesian single-subject analysis
• Assumption: posterior distribution of the parameters is gaussian
• Use of the cumulative normal distribution to test the probability by
which a certain parameter (or contrast of parameters cT ηθ|y) is
above a chosen threshold γ:
ηθ|y
Probability
ηθ|y
• γ can be chosen as zero ("does the effect exist?") or as a function
of the expected half life τ of the neural process: γ = ln 2 / τ
Group analysis
• In analogy to “random effects” analyses in SPM, 2nd level analyses
can be applied to DCM parameters:
Separate fitting of identical models
for each subject
Selection of bilinear parameters of
interest
one-sample t-test:
parameter > 0 ?
paired t-test:
parameter 1 >
parameter 2 ?
rmANOVA:
e.g. in case of multiple
sessions per subject
Model comparison and selection
Given competing hypotheses
on structure & functional
mechanisms of a system, which
model is the best?
Which model represents the
best balance between model
fit and model complexity?
For which model i does
p(y|mi) become maximal?
Pitt & Miyung (2002), TICS
Bayesian Model Selection
Bayes theorem:
Model evidence:
The log model
evidence can be
represented as:
Bayes factor:
p( y | , m) p( | m)
p( | y, m) =
p ( y | m)
p( y | m) = p( y | , m) p( | m) d
log p ( y | m) = accuracy (m) −
complexity(m)
p( y | m = i)
Bij =
p( y | m = j )
Penny et al. 2004, NeuroImage
The DCM cycle
Hypothesis about
a neural system
Statistical test
on parameters
of optimal model
Definition of
DCMs as system
models
Bayesian model
selection of
optimal DCM
Design a study that
allows to investigate
that system
Data acquisition
Parameter estimation
for all DCMs considered
Extraction of
time series
from SPMs
Inference about DCM parameters:
Bayesian fixed-effects group analysis
Because the likelihood
distributions from different
subjects are independent, one
can combine their posterior
densities by using the posterior
of one subject as the prior for the
next:
p ( | y1 )
p ( y1 | ) p ( )
p ( | y1 , y2 ) p( y2 | ) p ( y1 | ) p ( )
p( y2 | ) p( | y1 )
...
p ( | y1 ,..., y N ) p( y N | ) p( | y N −1 )... p( | y1 )
See:
spm_dcm_average.m
Neumann & Lohmann, NeuroImage 2003
Under Gaussian assumptions
this is easy to compute:
group
posterior
covariance
individual
posterior
covariances
N
C−|1y1 ,..., y N = C−|1yi
i =1
| y ,..., y
1
group
posterior
mean
N
N −1
−1
= C | yi | yi C | y1 ,..., y N
i =1
individual posterior
covariances and means
Approximations to model evidence
Laplace approximation:
F = accuracy (m) − complexity(m)
1
1
= − log Ce − (y − h(θ))T Ce−1 (y − h(θ))
2
2
1
1
1
− (θ | y − θ p )T C−p1 (θ | y − θ p ) − log C p + log C | y
2
2
2
Akaike information
criterion (AIC):
Bayesian information
criterion (BIC):
Unfortunately, the complexity
term depends on the prior
density, which is determined
individually for each model to
ensure stability. Therefore, we
need other approximations to
the model evidence.
AIC ( y | m) = accuracy (m) − p
p
BIC ( y | m) = accuracy (m) − log N S
2
Penny et al. 2004, NeuroImage
DCM parameters = rate constants
Integration of a first order linear differential equation gives
an exponential function:
dz
= az
dt
z (t ) = z0 exp( at )
The coupling parameter a is
inversely proportional to the
half life of z(t):
z ( ) = 0.5 z0
0.5 z0
The coupling parameter a
thus describes the speed of
the exponential growth/decay:
= z0 exp( a )
a = ln 2 /
= ln 2 / a