CHAPTER 6 EXPRESSION OF BIOLOGICAL INFORMATION SB015 2
advertisement

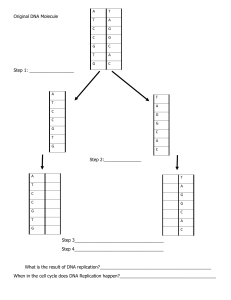
: CHAPTER 6 : EXPRESSION OF BIOLOGICAL INFORMATION (2L + 8T) 6.0 Expression of Biological Information 6.1 6.2 6.3 6.4 DNA & Genetic Information DNA Replication Protein Synthesis: Transcription & Translation Gene Regulation & Expression Learning outcomes 6.1 DNA & Genetic Information a) State the concept of Central Dogma. 6.2 DNA Replication a) Explain semi-conservative replication of DNA. b) Explain the enzymes and proteins that involve in DNA replication. c) Explain the mechanism of DNA replication. Learning Outcomes : Learning Outcomes : 6.1 (a) State the concept of Central Dogma 6.1 a) Explain DNA as the carrier of genetic information LECTURE Central Dogma ⮚ Dogma: a belief that is accepted without doubt or question ⮚ Central Dogma of molecular biology explain the flow of genetic information within biological system ⮚ Genetic information flows from: i. DNA to DNA (during replication before a cell divides) ii. DNA to protein (during its phenotypic expression in an organism) transcription DNA RNA reverse transcription replication translation protein Learning Outcomes : Learning Outcomes : 6.1 (a) State the concept of Central Dogma 6.1 a) Explain DNA as the carrier of genetic information LECTURE Central Dogma ⮚ Genetic information which flows from DNA to protein involves 2 steps: i. Transcription, the transfer of information from DNA to RNA ii. Translation, the transfer of information from RNA to protein transcription DNA RNA reverse transcription replication translation protein Learning Outcomes : Learning Outcomes : 6.1 (a) State the concept of Central Dogma 6.1 a) Explain DNA as the carrier of genetic information LECTURE Central Dogma ⮚ Genetic information also flows from RNA to DNA during the conversion of genomes of viral RNA to their DNA proviral forms ⮚ The transfer of genetic information from DNA to RNA is sometimes reversible ⮚ But the transfer of genetic information from RNA to protein is irreversible ⮚ The transfer of protein to protein or from protein to nucleic acid (DNA / RNA) also are not possible transcription DNA RNA reverse transcription replication translation protein Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA LECTURE DNA Replication • • • DNA is copied precisely before cell division → replication During S phase (interphase) 1 chromosome consists of 2 sister chromatids bound to a centromere Sister chromatids DNA replication Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA LECTURE DNA Replication • • 3 models of replication Meselson & Stahl (1957) suspected that DNA replication is semi-conservative DNA Replication Models Conservative Semi-conservative Dispersive Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA LECTURE Model 1 : Conservative Double stranded DNA Conservative Model *Note: • Blue (parental strand) • Red (new strand) • 2 parental strands may remain together after acting as templates • 2 newly synthesized strand form the second double helix Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA LECTURE Model 2 : Semi-conservative Double stranded DNA Semi-conservative Model *Note: • Blue (parental strand) • Red (new strand) • Both DNA molecules consist of 1 parental strand and 1 newly synthesized strand Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA LECTURE Model 3 : Dispersive Double stranded DNA Dispersive Model *Note: • Blue (parental strand) • Red (new strand) • Each strand of double stranded DNA contains a mixture of parental and new strands Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA LECTURE DNA Replication Conservative Model • Parental strand complements with parental strand • Newly synthesised strand complements with new strand Semi-conservative Model • Parental strand complements with new strand Dispersive Model • Each strand of double stranded DNA has combination of parental & new strands Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) • Use bacteria ~ Escherichia coli • Use 2 nitrogen isotopes: 1) 14N (normal light isotope of nitrogen) 2) 15N (heavy isotope of nitrogen) Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) • • • Nitrogen isotopes are used to form NH4Cl (added in the medium culture) Bacteria uses nitrogen to synthesize nitrogenous bases Which is used to synthesize DNA during replication before a cell divides Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) • 2 isotopes are used to label & differentiate parental strand DNA & newly synthesized DNA Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) • • E. coli is grown in a medium containing 15NH4Cl for several generations Some bacteria are transferred to a medium containing 14N Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) Escherichia coli Cultured in medium 15N F1 generation Transfer • • • E. coli is grown in a medium containing 15NH4Cl for several generations. Some bacterial samples were taken (F0). The rest are transferred to a medium containing 14N. ASSIGNMENT Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA Meselson & Stahl Experiment (1957) Cultured in medium 15N Transfer • • • F1 generation F1 generation Extract DNA Centrifuge After one generation, some bacteria is isolated (F1 generation) while others continue to grow in 14N medium DNA is extracted from the bacterial sample (F1) Purified DNA is subjected to density gradient centrifugation Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) Cultured in medium 15N F1 generation Extract DNA F2 generation Transfer F3 generation • • Bacteria in medium 14N is allowed to replicate twice to produce F2 generations & thrice (F3 generations) Repeat DNA extraction & centrifugation steps for F2 and F3 generations ASSIGNMENT Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA Meselson & Stahl Experiment (1957) E. coli cultured in medium 15N Transfer to medium 14N DNA extraction Generation F0 One cell division DNA extraction Generation F1 DNA extraction Generation F2 2 cell divisions 3 cell divisions DNA extraction Generation F3 Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT ASSIGNMENT Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA Meselson & Stahl Experiment (1957) F0 generation Control 14N14N 15N15N • Molecules of different density can be separated by density gradient centrifugation • DNA of cells grown in medium containing 14N have a lower density than DNA of cells grown in 15N • Parental DNA (generation F0) accumulates in high-density region Model 1 : Conservative ASSIGNMENT 14N14N 14N14N 14N14N 14N14N 14N14N 14N14N 14N14N 14N14N 15N15N 14N14N 14N14N 15N15N 14N14N 15N15N 15N15N Parental / F0 generation F1 generation F2 generation F3 generation 14N14N Hybrid DNA 14N/15N 15N15N Model 2 : Semi-conservative ASSIGNMENT 14N14N 14N14N 14N14N 14N15N 14N15N 14N15N 14N14N 15N15N 14N14N 14N15N F1 generation 14N14N 14N14N 14N15N Parental / F0 generation 14N14N F2 generation 14N15N F3 generation 14N14N Hybrid DNA 14N/15N 15N15N ASSIGNMENT Model 3 : Dispersive 14N15N 14N15N 14N15N 14N15N 14N15N 14N15N 14N15N 15N15N 14N15N 14N15N F1 generation 14N15N 14N15N 14N15N Parental / F0 generation 14N15N F2 generation 14N15N F3 generation 14N14N Hybrid DNA 14N/15N 15N15N Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment - Results RESULT 14N14N Hybrid DNA 14N/15N 15N15N 14N14N Hybrid DNA 14N/15N 15N15N 14N14N Hybrid DNA 15N15N 14N/15N ASSIGNMENT Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA Meselson & Stahl Experiment (1957) 14N14N F0 generation Control 14N14N 15N15N 15N15N • Molecules of different density can be separated by density gradient centrifugation • DNA of cells grown in medium containing 14N have a lower density than DNA of cells grown in 15N • Parental DNA (generation F0) accumulates in high-density region Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment (1957) FO generation F1 generation F2 generation F3 generation ASSIGNMENT Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment - Results Parental / F0 generation F1 generation F2 generation F3 generation 14N14N Hybrid DNA 14N/15N 15N15N • • • For F1 generation, 1 band is obtained (have intermediate density known as hybrid density) For F2 generations, 2 bands exist. 1 band is hybrid density & the other band was light After 3 generations, 2 bands exist: intensity of ¼ hybrid, ¾ light band Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA ASSIGNMENT Meselson & Stahl Experiment - Results • • • For F1 generation, only 1 intermediate density band is obtained (between the density of light & heavy DNA a.k.a hybrid density) For F2 generations, 2 bands exist. 1 band is hybrid density & the other band was light After 3 generations, DNA band was ¼ hybrid, ¾ light band Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA Meselson & Stahl Experiment Conclusion: • DNA replicates by semi-conservative ASSIGNMENT ASSIGNMENT Learning Outcomes : 6.2 (a) Explain semi-conservative replication of DNA DNA Replication Hydrogen bonds formed between complementary bases DNA is a double helix of two polynucleotide strands Complementary base pairs hold the two DNA35strands together Learning Outcomes Outcomes :: Learning 6.2(c) (c) Explain Explain the of DNA replication and the enzymes 4.1 the mechanism Mendel’s experiments on monohybrid cross involved DNA Replication • The process involves are : Step 1: Separation of DNA double helix Step 2: Building RNA primer Step 3: DNA synthesis Step 4: Replacing RNA primer Step 5: Joining of Okazaki fragments (ligation) Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved • Initiator protein binds to origin of replication • Helicase unwind the DNA double helix Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved • DNA replication is bidirectional from the origin of replication (but will consider events at just one replication fork) Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 1: Separation of DNA double helix 5’ G A T C C G T T 3’ A A C C G T A A T C T G C T A T C G G A C A T C T T T G G C A T T A G AC G A T A G C C T G T A G A C T A G G C A A 5’ Replication fork 3’ • Helicase unwind DNA double helix & break the hydrogen bond between complementary nitrogenous bases • 2 polynucleotide strands separate • Each strand act as a template for the synthesis of a new strand Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved SSB Proteins 5’ G A T C C G T T 3’ A A C C G T A A T C T G C T A T C G G A C A T C T T T G G C A T T A G AC G A T A G C C T G T A G A C T A G G C A A 3’ 5’ Replication fork • Single-strand binding proteins (SSB proteins) bind to the separated DNA strand to prevent them from rejoining Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved overwinding Overwinding supercoil supercoil • When double stranded DNA is unwound, it adds extra helical twist beyond the unwound region • DNA become more tightly twisted (overwinding/supercoil), creating a strain to DNA strand Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved overwinding Overwinding supercoil topoisomerase nick supercoil • Topoisomerase makes a nick by breaking the phosphodiester bond • Topoisomerase helps relieve the strain of unwound double stranded DNA and rejoin the DNA strands by reforming phosphodiester bond Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 1: Separation of DNA double helix Step 2: Building RNA primer U C G U G A A A C G A T RNA nucleotide G C G G A T C Primase 5’ T A T DNA nucleotide DNA polymerase III G A T C C G T T 3’ A A C C G T A A T C T G C T A T C G G A C A T C T 5’ T T G G C A T T A G AC G A T A G C C T G T A G A G A U 5’ C T A G G C A A Replication fork 3’ G A U C A G C G A T A T C A C G A T T Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 2: Building RNA primer • DNA polymerase III cannot begin the synthesis of a new strand • It can only extend an existing strand paired with a template strand • To begin synthesis, a short fragment of RNA, called RNA primer, must be created and paired with the template DNA strand Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 2: Building RNA primer • Each strand act as a template • Primase catalyze the synthesis of a short RNA primer that are complementary to the template DNA strand • RNA primer is about 10-60 nucleotides long • Elongation of a new strand is from 5’ to 3’ Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 2: Building RNA primer • RNA primer is synthesized to provide free 3’-hydroxyls end • Which allow DNA polymerase III to start the synthesis of DNA strand • by adding DNA nucleotide to the free 3’-hydroxyls end of these RNA primers Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 2: Building RNA primer U C G U G A A C A G A T RNA nucleotide G C T G A T C Primase G A T DNA nucleotide 5’ DNA polymerase III G A T C C G T T 3’ A A C C G T A A T C T G C T A T C G G A C A T C T 5’ T T G G C A T T A G AC G A T A G C C T G T A G A G AUC C G T T 5’ C T A G G C A A Replication fork 3’ G A U C A G C G A T A T C A C G A T T Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 3: DNA synthesis • • DNA polymerase III catalyze the synthesis of new DNA strand by adding DNA nucleotides to the short RNA primer The adding of DNA nucleotides is complementary to the bases of the template Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 3: DNA synthesis Primase 5’ DNA polymerase III G A T C C G T T 3’ C T A G G C A A 3’ 5’ 5’ T T G G C A T T A G AC G A T A G C C T G T A G A G AUC C G T T C T A G G C A A 3’ A A C C G T A A T C T G C T A T C G G A C A T C T 5’ Replication fork Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 3: DNA synthesis Primase 5’ DNA polymerase III G A T C C G T T 3’ C T A G G C A A 3’ 5’ 5’ T T G G C A T T A G AC G A T A G C C T G T A G A G AUC C G T T C T A G G C A A 3’ A A C C G T A A T C T G C T A T C G G A C A T C T 5’ Replication fork Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 3: DNA synthesis 5’ G A T C C G T T A G G A A A C C G T A A T C 3’ C T A G G C A A T C C T T T G G C AT UA G 3’ 5’ 5’ G AUC C G T T A G G A A AC C G T AA T C C T A G G C A A T C C T T T G G C A T T A G 3’ Primase T C G G A C A T C T A G C C T G T A G A 5’ Replication fork DNA polymerase III Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 3: DNA synthesis 5’ 3’ G A T C C G T T A G G A A A C C G T A A T C T G C T A T C G G A C A T C T C T A G G C A A T C C T T T G G C A T U A G AC G A T A G C C T G T A G A 3’ 5’ G A T C C G T T A G G A A A C C G T A A T C T G C T A T C G G A C A T C T 5’ 3’ C T A G G C A A T C CT T T G G C A T T A G AC G A T A G C C T G T A G A 3’ 5’ Primase DNA polymerase III Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 3: DNA synthesis • • • DNA is synthesized continuously / towards the replication fork is called as the leading strand On another strand, DNA is synthesized discontinuously / away from the replication fork is called as the lagging strand Producing short Okazaki fragments • DNA polymerase I replace RNA primer with DNA nucleotides Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 4: Replacing RNA primer DNA polymerase I 5’ 3’ G A T C C G T T A G G A A A C C G T A A T C T G C T A T C G G A C A T C T C T A G G CC A AA A T C C T T T G G C AT U TA A GG A C G A T A G C C T G T A G A 3’ 5’ G A T C C G T T A G G A A A C C G T A A T C T G C T A T C G G A C A T C T 5’ 3’ C T A G G C A A T C CT T T G G C A T T A G AC G A T A G C C T G T A G A 3’ • 5’ DNA polymerase I replace RNA primer with DNA nucleotides Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Step 5: Joining of Okazaki fragments (ligation) • DNA ligase joins Okazaki fragments • By catalyzing the formation of phosphodiester bond between adjacent Okazaki fragments • 2 identical copies of DNA are produced DNA ligase 5’ 3’ G A T C C G T T A G G A A A C C G T A A T C T G C T A T C G G A C A T C T C T A G G C A A T C C T T T G G C A T T A G AC G A T A G C C T G T A G A 3’ 5’ G A T C C G T T AG G A AA C C G T A A T C T G C T A T C G G A C A T C T 5’ 3’ C T A G G C A A T C CT T T G G C A T T A G AC G A T A G C C T G T A G A 3’ 5’ Learning Outcomes Outcomes :: Learning 6.2(c) (c) Explain Explain the of DNA replication and the enzymes 4.1 the mechanism Mendel’s experiments on monohybrid cross involved DNA Replication • The process involves are : Step 1: Separation of DNA double helix Step 2: Building RNA primer Step 3: DNA synthesis Step 4: Replacing RNA primer Step 5: Joining of Okazaki fragments (ligation) Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved • Initiator protein binds to origin of replication • Helicase unwind the DNA double helix Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved 5’ 3’ Learning Outcomes : 6.2 (c) Explain the mechanism of DNA replication and the enzymes involved Learning Outcomes : 6.2 (b) Explain the enzymes and protein involved in DNA replication Function of Enzymes & Proteins in DNA Replication Enzymes or Protein Function 1. Helicase Unwind the parental DNA double helix and separate DNA strands by breaking hydrogen bonds between complementary bases 2. Single Strand Binding Proteins (SSB protein) Binds to the separated DNA strands to prevent them from rejoining 3. Topoisomerase Relieve the strain of unwound DNA by breaking, swiveling and rejoining DNA strands 4. Primase Catalyze the synthesis of a short RNA primer Learning Outcomes : 6.2 (b) Explain the enzymes and protein involved in DNA replication Function of Enzymes & Proteins in DNA Replication Enzymes 5. DNA polymerase III Function Catalyze the synthesize of new DNA strand by adding complementary DNA nucleotide to the 3’end of RNA primer (which must be complementary to the template) 6. DNA polymerase I 7. DNA ligase Replace RNA primers with DNA nucleotides Catalyze the formation of phosphodiester bond between Okazaki fragments Learning outcomes 6.3 Protein Synthesis: Transcription & Translation a) Explain briefly transcription and translation. b) Introduce codon & its relationship with sequence of amino acid using genetic code table. a) Explain transcription and the stages involved in the formation of mRNA strand from 5’ to 3’. a) Explain translation and the stages involved. Learning Outcomes : Learning Outcomes : 6.3 a) Explain briefly transcription and translation 6.1 a) Explain DNA as the carrier of genetic information LECTURE Central Dogma • Genetic information flows from DNA to protein involves 2 steps: i. Transcription, the transfer of information from DNA to RNA ii. Translation, the transfer of information from RNA to protein • This flow of genetic information a.k.a as gene expression transcription DNA RNA reverse transcription replication translation protein Learning Outcomes : 6.3 a) Explain briefly transcription and translation LECTURE Protein Synthesis 1. Transcription • synthesise mRNA A process by which genetic information in DNA base sequence is copied or transcribed into complementary sequence of mRNA. 2. Translation • synthesise protein The sequence of nucleotides in mRNA act as a template and is translated into sequence of amino acids. Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table LECTURE Properties of Genetic Code Functional mRNA 5’ 3’ A U G C A U U U G C GA G A C U A A 1st Codon • • 2nd Codon 3rd Codon 4th Codon 5th Codon 6th Codon 3 base sequence of mRNA (triplet code) is called codon 1 codon on mRNA codes for 1 amino acid Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table LECTURE 1st base (5’ end) U 2nd base (5’ end) U UUU UUC UUA UUG C Phe Leu UCU UCC UCA UCG C CUU CUC CUA CUG CCU CCC Leu CCA CCG A AUU AUC AUA AUG ACU ACC ACA ACG G GU U GU C GU A GU G Ile Met/star t Val GC U GC C GC A GC G A Ser Pro Thr Ala 3rd base (5’ end) G UAU Tyr UAC U A A STO STO UAG P P CAU His CAC CAA Gln CAG U GU Cys U GC U G A STO U G G P Trp U C A G C GU C GC C GA C GG U C A G AAU AAC AAA AAG A GU A GC A GA A GG GA U GA C GA A GA G Asn Lys Asp Glu GGU GGC GGA GGG Arg Ser Arg Gly U C A G U C A G Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 1. Composed of nucleotide triplets (triplet code) • 3 nucleotides in mRNA is called a codon • If 2 nucleotides per codon • will result in only 42 or 16 possible codon (rejected) LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 1. Composed of nucleotide triplets (triplet code) • • 3 bases per codon will result in 43 or 64 possible codons LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 2. Each codon represents 1 specific amino acid • 20 different amino acids commonly found in proteins • At least 20 different codons LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 3. The code is degenerate • Most amino acids are coded by more than one codon • Only 2 amino acids are coded by 1 codon • 61 codons represent 20 amino acids LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 4. Contain start & stop codons • AUG is the start codon. • UAG, UAA, UGA are the stop codons. • Do not code for any amino acid. • Signals to terminate the coding sequence. LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 5. Almost universal • The same codon codes for the same amino acids in all organisms LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 6. Non-overlapping • • • • Each codon consists of 3 nucleotides Sequence of codon is read continuously from a fixed starting point & proceed until it reaches the termination point . LECTURE Learning Outcomes : 6.3 c) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 7. Comma-free (commaless) • • • • There are no commas or other forms of punctuation within the coding regions of mRNA Codons are read consecutively LECTURE Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand Transcription • • • Occurs in nucleus (for eukaryote). Involves RNA polymerase enzyme. During transcription, one DNA strand acts as a template to synthesise a complementary strand of mRNA. ASSIGNMENT Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand Transcription • The processes involved are : Step 1: Initiation Step 2: Elongation Step 3: Termination ASSIGNMENT Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Transcription • • • DNA consists of many genes. Each gene has a promoter, coding region and a terminator. Promoter: ✔ marks the beginning of a gene ✔ determines which strands of DNA is used as a template • Coding region: base triplet that codes for amino acid • Terminator: marks the end of a gene Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Stage 1 : Initiation • • • RNA polymerase binds to promoter on DNA. It moves along the DNA in the direction of transcription. It catalyses the unwinding of the double helix DNA. Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Stage 1 : Initiation • • • RNA polymerase catalyses the breaking of hydrogen bonds between complementary nitrogenous bases. Both DNA strands are separated. One of DNA strand is used as a template for transcription. Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Stage 2 : Elongation • • RNA polymerase catalyses addition of nucleotides in the direction of 5’ to 3’ end. RNA polymerase catalyses addition of free ribonucleotides which are complementary to the bases on the DNA template strand. Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Stage 2 : Elongation • • Uracil pairs with Adenine (on DNA). Cytosine pairs with Guanine. DNA-RNA hybrid is temporarily formed. Stage 2 : Elongation • • ASSIGNMENT Newly synthesised mRNA (upstream) detaches from DNA template. DNA double helix re-forms / rewinds, leaving only few DNA-RNA hybrid regions. ASSIGNMENT Transcription: Stage 2 (Elongation) Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Stage 3 : Termination • • • • Elongation proceeds until RNA polymerase reaches termination site. Terminator sequence gives signal to RNA polymerase to stop adding nucleotides to the mRNA strand. RNA polymerase detaches. mRNA molecule is released. Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Stage 3 : Termination • • • • • RNA polymerase detaches. mRNA molecule is released from DNA template. Double stranded DNA rejoins to form double helix again. In prokaryote, mRNA doesn’t need any modification. In eukaryote, further modification is needed; thus after transcription, its mRNA is called as pre-mRNA. Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand Transcription A single gene can be transcribed simultaneously by several molecules of RNA polymerase, • increases the number of mRNA molecules • allows a cell to produce specific protein in large amount ASSIGNMENT Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT Differences between DNA Replication & Transcription DNA Replication Transcription Both DNA strands act as template Only one strand act as a template Whole DNA genome is copied Only a small part of DNA genome is copied Produce two molecules of (double Produce a single stranded mRNA stranded) DNA strand Nitrogenous bases used is guanine, adenine, cytosine and thymine Nitrogenous bases used is guanine, adenine, cytosine and uracil RNA primer is required RNA primer is not required Enzyme involved is DNA polymerase Enzyme involved is RNA polymerase Only occurs during S phase of interphase Occurs throughout the life of a cell Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT mRNA Modification (RNA Splicing) in Eukaryotes • • • In eukaryotes, both coding sequences (exons) and noncoding sequences (introns) are found along a DNA. Coding sequences are not continuous. Introns are found between coding regions (exons). Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand ASSIGNMENT mRNA Modification (RNA Splicing) in Eukaryotes • • Pre-mRNA contains exon & intron regions. Process of removing the introns and recombining all the coding exon regions is RNA splicing (in nucleus). Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand RNA Splicing in Eukaryotes • • • In RNA splicing, the introns are cut out from pre-mRNA by spliceosome Spliceosome is made up of proteins and small RNA Spliceosome binds to specific nucleotide sequence along an intron ASSIGNMENT Learning Outcomes : 6.3 c) Explain transcription and the stages involved in the formation of mRNA strand RNA Splicing in Eukaryotes • • • • The intron is cut out and released. The exons that flanked the intron together are joined. Functional mRNA is produced. mRNA moves from nucleus into cytoplasm (ribosome) to be translated. ASSIGNMENT LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code Functional mRNA 5’ 3’ A U G C A U U U G C GA G A C U A A 1st Codon • • 2nd Codon 3rd Codon 4th Codon 5th Codon 6th Codon 3-base sequence of mRNA (triplet code) is called codon. 1 codon on mRNA codes for 1 amino acid. LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table 1st base (5’ end) U 2nd base (5’ end) U UUU UUC UUA UUG C Phe Leu UCU UCC UCA UCG C CUU CUC CUA CUG CCU CCC Leu CCA CCG A AUU AUC AUA AUG ACU ACC ACA ACG G GU U GU C GU A GU G Ile Met/star t Val GC U GC C GC A GC G A Ser Pro Thr Ala 3rd base (5’ end) G UAU Tyr UAC U A A STO STO UAG P P CAU His CAC CAA Gln CAG U GU Cys U GC U G A STO U G G P Trp U C A G C GU C GC C GA C GG U C A G AAU AAC AAA AAG A GU A GC A GA A GG GA U GA C GA A GA G Asn Lys Asp Glu GGU GGC GGA GGG Arg Ser Arg Gly U C A G U C A G LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 1. Composed of nucleotide triplets (triplet code) • • • 3 nucleotides in mRNA is called a codon If 2 nucleotides per codon will result in only 42 or 16 possible codon (rejected) 3 bases per codon will result in 43 or 64 possible codons LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 2. Each codon represents 1 specific amino acid • 20 different amino acids commonly found in proteins. • At least 20 different codons. 3. The code is degenerate • Most amino acids are coded by more than one codon. • Only 2 amino acids are coded by 1 codon. • 61 codons represent 20 amino acids. LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 4. Contain start & stop codons • AUG is the start codon. • UAG, UAA, UGA are the stop codons. • Signals to terminate the coding sequence (do not code for any amino acid). 5. Almost universal • With few exceptions, the same codon codes for the same amino acids in all organisms. LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 6. Non-overlapping • Each codon consists of 3 nucleotides and the following codons are represented by the following triplets. • Sequence of codon is read continuously from a fixed starting point & proceed until it reaches the termination point at the other end LECTURE Learning Outcomes : 6.3 b) Introduce codon and its relationship with sequence of amino acid using genetic code table Properties of Genetic Code 7. Comma-free (commaless) • • There are no commas, or other forms of punctuation within the coding regions of mRNA. Codons are read consecutively. QUESTION • • • • • What is the base sequence of the Codon 1? [1 mark] What is the anticodon sequence for codon 1? [1 mark] Name the enzyme involved in the production of Y. [1 mark] Name X and give its function in protein synthesis. [2 marks] Give THREE differences between molecules X and Y. [3 marks] Reference • Campbell N.A et. al., Biology, 11th ed. (2018), Pearson Education Limited. (page 392-397) • Snustad D. P., Simmons M. J and Jenkins, J. B Principles of Genetics, (1997), John Wiley & Sons, Inc, (page 297-299, 303-305) Learning Outcomes : 6.3 d) Explain translation and the stages involved LECTURE Protein Synthesis 1. Transcription • synthesise mRNA Process by which genetic information in DNA base sequence is copied or transcribed into complementary sequence of mRNA 2. Translation • synthesise protein The sequence of nucleotides in mRNA act as a template and is translated into sequence of amino acids Learning Outcomes : 6.3 d) Explain translation and the stages involved LECTURE Types of RNA a) mRNA (Messenger RNA) • single stranded RNA • carry genetic information transcribed from DNA in nucleus to ribosomes for translation in cytoplasm. • The triplet bases on the mRNA are called codon Learning Outcomes : 6.3 d) Explain translation and the stages involved Types of RNA b) rRNA (Ribosomal RNA) • Major component of ribosome • Combines with protein to form ribosomes LECTURE Learning Outcomes : 6.3 d) Explain translation and the stages involved Ribosome • • • • Site of protein synthesis Has 2 subunits Small subunits – contain mRNA binding site Large subunits – contain 3 tRNA binding site i.e: ⮚ E site ⮚ P site ⮚ A site LECTURE Learning Outcomes : 6.3 d) Explain translation and the stages involved LECTURE Ribosome • • • E site (exit site) P site (peptidyl-tRNA binding site) A site (aminoacyl-tRNA binding site) E Large subunit Small subunit P site A site P A Learning Outcomes : 6.3 d) Explain translation and the stages involved Types of RNA c) tRNA (Transfer RNA) • • • • • Single strand RNA (~70-80 nucleotides long). It folds back on itself to form a compact 3D shaped. Forms a cloverleaf shape, held by hydrogen bonds between complementary bases. The middle loop contains 3 bases called anticodon. The protruding 3’ end (5’ CCA 3’ ending) act as attachment site of a specific amino acid. LECTURE Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Activation of Amino Acids • • • mRNA codon carries the code for amino acids. Anticodon (of tRNA) has a specific complementary binding sequence for the codon (of mRNA). tRNA that binds to the codon in mRNA must carry the amino acid encoded by that codon to the ribosome to ensure accurate translation. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Activation of Amino Acids • • tRNA will bind with the amino acid that matches the code for the correct mRNA codon. Each kind of tRNA binds to a specific amino acid. Learning Outcomes : 6.3 d) Explain translation and the stages involved Activation of Amino Acids Activation is a process in which a specific amino acid is attached to a tRNA molecule • to form aminoacyl-tRNA, • catalysed by aminoacyl-tRNA synthetase. • ATP is used as energy source. • There are 20 different aminoacyl-tRNA synthetases. • tRNA is re-activated many times. ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved Activation of Amino Acids • • Amino acids are linked to their respective tRNA molecules by the formation of ester bond, at the 3’end of tRNA. ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved Activation of Amino Acids ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved Translation • The process involves are : Step 1: Initiation Step 2: Elongation Step 3: Termination ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 1 : Initiation • • • • Small ribosomal subunit binds to mRNA. It binds to the specific nucleotide sequence just upstream of the start codon, AUG (in bacterial cell). Initiator aminoacyl-tRNA that carries methionine (met) binds to the start codon (held by hydrogen bonds). The sequence of start codon (5’AUG3’) is complementary with anticodon (3’UAC5’) found in initiator tRNA. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 1 : Initiation • • • • Then, large ribosomal subunit attaches with the initiation complex Completing the translation initiation complex. Protein called initiation factors are needed to assemble all these components together. Energy is provided by the hydrolysis of GTP. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 1 : Initiation • • • Initiator tRNA is located in the P site of the ribosome. A site is empty. Available for the next aminoacyl-tRNA to enter (that has a complementary sequence of anticodon to the second codon on mRNA). Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 2 : Elongation • In the elongation stage, amino acids are added one by one to the previous amino acid at the C-terminus of the growing chain.. Learning Outcomes : 6.3 d) Explain translation and the stages involved Step 2 : Elongation • Each addition involves a 3-step cycle:1) Codon recognition 2) Peptide bond formation 3) Translocation • Energy is supplied by hydrolysis of GTP in steps 1 & 3 ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 2 : Elongation • During codon recognition, aminoacyl-tRNA with anticodon which is complementary to the mRNA codon enters to the empty A site. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 2 : Elongation • Peptide bond is formed between the carboxyl group of amino acid in the P site & the amino group of the new amino acid in A site. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 2 : Elongation • • Catalysed by peptidyl transferase, a large subunit rRNA that acts as enzyme, thus called ribozyme. This step removes the growing polypeptide from the tRNA in the P site and binds it to the new amino acid on the tRNA in the A site by a peptide bond. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 2 : Elongation • • Ribosome moves a codon ahead from 5’ to 3’ direction. tRNA in the A site is now translocated to the P site. • This is called translocation. Initiator-tRNA is now in E site, exits from ribosome. A site is now empty. • • Learning Outcomes : 6.3 d) Explain translation and the stages involved Step 2 : Elongation • • • • The 3rd aminoacyl-tRNA enters A site. Peptide bond is formed between 2nd & 3rd amino acids. The growing polypeptide chain is released from tRNA in the P site, and binds to the amino acid attached to the tRNA in the A site by a peptide bond. ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved Step 2 : Elongation ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 2 : Elongation • • Amino acids are added one by one to the previous amino acid. The whole process is repeated until all the codons of mRNA is translated. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 3 : Termination • • • • Elongation continues until a stop codon in the mRNA reaches the A site. The stop codons (UAG, UAA, UGA) do not code for amino acids; thus no tRNA will bind to a stop codon. Stop codon acts as a signal to stop translation. A release factor - a protein that recognises the stop codon binds to the stop codon in the A site. Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Step 3 : Termination • • • • The release factor causes the addition of water molecule to the polypeptide chain. This reaction hydrolyses the bond between the last amino acid of the polypeptide chain and the tRNA in the P site. Newly synthesised protein is released. Ribosomal subunits dissociates. Learning Outcomes : 6.3 d) Explain translation and the stages involved Protein Synthesis ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved Polyribosomes Structure ASSIGNMENT Learning Outcomes : 6.3 d) Explain translation and the stages involved ASSIGNMENT Polyribosomes Structure • • • A single mRNA strand is used to make many copies of polypeptide. Multiple ribosomes may attach on the same mRNA at a time. It forms a structure called polyribosomes / polysome. QUESTION • Name the stage of translation process shown in the above figure. [1 mark] • Which is the first codon used in protein synthesis from this mRNA? [1 mark] • What is the anti-codon sequence in tRNA 1? [1 mark] QUESTION • tRNA 1 has a 5’ – phosphate end and a 3’ – hydroxyl end. What is the function of 3’OH end in tRNA 1? [1 mark] • Name the enzyme that catalyzes the formation of the peptide bond between amino acids carried by tRNA 1 and tRNA 2. [1 mark] Learning outcomes 6.4 Gene Regulation & Expression: lac operon a) Explain the concept of operon and gene regulation. b) State the components of operon. c) Explain the components of lac operon and their functions in E. coli. a) Explain the mechanism of the operon in the absence and presence of lactose. Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation LECTURE lac Operon • • • Operon model describes the mechanism for the control of gene expression in bacteria. Some of the bacterial genes will be expressed only when it is necessary. The operon model was proposed by Jacob and Monod (in 1961) on E. coli Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation LECTURE Concept of Operon What is operon? • Operon is a complete unit of gene expression in prokaryote. • Operon consist of a group of two or more structural genes having a related function. • Plus DNA sequences responsible for controlling them. Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation LECTURE Concept of Operon • • • Operon is regulated as a single transcriptional unit by a single promoter. These genes are transcribed into a single mRNA. To allow bacteria to synthesize all enzymes needed to metabolise a substrate at once if the substrate is present. Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation LECTURE Concept of Operon • 1) 2) 3) • Operon consists of: Promoter Operator Structural genes Operon is controlled by a regulatory gene (upstream from the operon). Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation ASSIGNMENT Gene Regulation & Expression (lac Operon) • • • Francois Jacob & Jacques Monod (1961) conduct an experiment by using E. coli E. coli lives in the human’s colon & depends on glucose or lactose consumed by human. When both glucose & lactose are present, E. coli prefer to metabolise glucose. Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation ASSIGNMENT Gene Regulation & Expression (lac Operon) • • • Until glucose is depleted; then only it uses lactose, but it does not grow immediately. After a short delay, it starts to use lactose as energy source & grows rapidly on the glucose medium. E. coli responds to changing environmental conditions. Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation ASSIGNMENT lac Operon • • • lac operon is a group of structural genes (3 genes) together with its promoter and operator, that involves in the metabolism of lactose which is controlled by regulatory gene In lac operon, the 3 genes (that codes for 3 enzymes) are controlled as a single unit on E. coli Allow E. coli to synthesize all 3 enzymes needed to metabolise lactose rapidly. lac Operon Regulatory gene lacI Promoter Operator P O Structural genes lacZ lacY lacA Learning Outcomes : 6.4 a) Explain the concept of operon & gene regulation ASSIGNMENT lac Operon • • When lactose is absent → lac operon is inactivated. When lactose is present (glucose absent) → lac operon is activated → structural genes are transcribed. lac Operon Regulator lacI Promoter Operator P O Structural genes lacZ lacY lacA Learning Outcomes : 6.4 b) State the components of lac operon LECTURE Components of lac Operon & Its Function • Operon consist of: 1) Promoter 2) Operator 3) Structural genes (lacZ, lacY & lacA) • Promoter & Operator are binding sites on DNA; they are not transcribed lac Operon Regulator lacI Promoter Operator P O Structural genes lacZ lacY lacA Learning Outcomes : 6.4 b) Explain the components of lac operon & their functions in E. coli ASSIGNMENT Components of lac Operon & Its Function • • • Structural genes are transcribed into a single mRNA. mRNA contains coding information for 3 enzymes & is translated to form 3 separate enzymes. Regulator gene carries the genetic code to produce repressor molecule that prevents the structural genes from becoming active. lac Operon Regulatory gene lacI Promoter Operator P O Structural genes lacZ lacY lacA Learning Outcomes : 6.4 b) Explain the components of lac operon & their functions in E. coli LECTURE Components of lac Operon & Its Function • Operon consists of: Gene Function 1. Regulatory gene lacI Gene that encodes for repressor protein 2. Promoter (P) Attachment site for RNA polymerase to start the transcription of structural genes 3. Operator (O) Attachment site of repressor protein 4. Structural genes lacZ Gene that encodes for β-galactosidase lac Y Gene that encodes for lactose permease lac A Gene that encodes for transacetylase Learning Outcomes : 6.4 b) Explain the components of lac operon & their functions in E. coli LECTURE Function of Proteins Expressed by lac Operon Protein Function 1. Repressor protein Controls whether the operon is activated or not 2. β-galactosidase Catalyses the hydrolysis of lactose into glucose & galactose 3. lactose permease Transports lactose into the cell efficiently (increase the permeability of cell membrane towards lactose) 4. transacetylase Transfers acetyl group from acetyl co-A to lactose Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Absence of Lactose (And Glucose Absent) lacI P Transcription 5’ 3’ Binding site for RNA polymerase O lacZ lacY Binding site for repressor Translation Repressor Protein • • • • Regulatory gene (lacI) is expressed continuously (constitutive). A repressor protein is produced. When lactose is absent, repressor protein is active. Repressor protein binds to operator. lacA Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Absence of Lactose (And Glucose Absent) Binding site for repressor lacI RNA polymerase • • • • P O lacZ lacY lacA Repressor Protein Repressor protein blocks RNA polymerase from transcribing the structural genes. Causes lac operon to be inactivated (“switched off”). Transcription of structural genes cannot occur. Enzymes needed to digest lactose cannot be produced. Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Presence of Lactose (Glucose Absent) Binding site for repressor lacI RNA polymerase • • • P O lacZ lacY lacA Repressor Protein When lactose is present, a few lactose enter the cells & converted to allolactose (isomer of lactose). Allolactose binds to the repressor protein. Thus, alter the shape of repressor protein (becomes inactive). Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Presence of Lactose (Glucose Absent) Binding site for repressor lacI P O lacZ lacY lacA RNA polymerase Repressor Protein • • • Repressor protein can no longer bind to the operator; it detaches from operator. RNA polymerase can transcribe the structural genes (transcription can occur). Allolactose acts as inducer that induces the transcription of lac operon. Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Presence of Lactose (Glucose Absent) Binding site for repressor lacI P O lacZ lacY RNA polymerase lacA Transcription 3’ 5’ • • • • Transcription of all 3 structural genes occur, to synthesize a single mRNA. Translation occurs to produce 3 enzymes: β-galactosidase, lactose permease & transacetylase. Translation β-galactosidase transacetylase lactose permease Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Presence of Lactose (Glucose Absent) Transcription occur Lac I P O lac Z lac Y lac A 3’ 5’ Translation β-galactosidase transacetylase lactose permease Learning Outcomes : 6.4 d) Explain the mechanism of lac operon in the absence and presence of lactose ASSIGNMENT Function of Enzymes • • • β-galactosidase → to hydrolyse lactose molecule into glucose & galactose Lactose permease → facilitate lactose to diffuse into the cell membrane Transacetylase → transfer acetyl group from acetyl coA to lactose ASSIGNMENT Reference • Neil A. Campbell, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky & Rebecca B. Orr (2021), Campbell Biology, 12th ed., Pearson Education Limited.(page 385-406) • Solomon E.P., Martin,C., Martin, D.W. & Berg, L.R. (2019), Biology, 11th ed. Cengage Learning Asia Pte Ltd. • Snustad D. P., Simmons M. J and Jenkins, J. B Principles of Genetics, (1997), John Wiley & Sons, Inc, (page 297-299, 303-305)