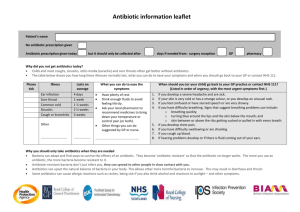
Fill-in Answer Sheet Instructions Before you start filling in the blanks, download this file and save it to your local drive or cloud. You can... ▶ personalize the file name. ▶ save the file when it is complete. ▶ attach it to your CarolinaScienceOnline assignment submission or an email. Prelab (continued) STUDENT GUIDE Name Date Prelab Questions 1. Read the Background and the Procedure. What question should you be able to answer using the data generated in this lab? 2. On days 2–7 you transfer some of each culture to a new tube with fresh LB broth and, for the culture that was grown in the presence of antibiotic, fresh ampicillin. Why in this experiment would it be critical to maintain the concentration of ampicillin? Question for deeper discussion: What might cause the level of ampicillin in the culture to decrease? 3. At the end of the experiment, what do you think the phenotype will be for the bacteria that are grown with ampicillin? Explain your answer. 4. At the end of the experiment, what do you think the phenotype will be for the bacteria that are grown without ampicillin? Explain your answer. ©2018 Carolina Biological Supply Company Fill-in • S-4 Evolution in Real Time: Bacteria and Antibiotic Resistance Laboratory Investigation (continued) STUDENT GUIDE Name Date Day 8: Assaying the Culture for Antibiotic Resistance Materials For each group: 2 cultures from lab day 7 sterile culture tubes, 6 LB plates, 2 LB/amp plates, 2 50-mL bottle sterile LB broth pipetting devices and sterile pipets for sterilely measuring and dispensing 0.9 mL and 1 mL of LB broth pipet and sterile tips for measuring 10 μL, 20 μL, and 100 μL culture tube rack benchtop waste container permanent marker Bunsen burner (may be shared between 2 groups) Shared: 37°C incubator spreading beads Reflection Questions •• Why do the cultures need to be diluted before they are plated? •• If you did not resuspend the culture before plating, how would that affect the results? Procedure Use sterile technique for all of the manipulations with bacteria and any plates or LB broth. When making serial dilutions refer to the diagram below to clarify the steps. 1. Label the six sterile culture tubes as follows: L, 1:10 L, 1:1000 L, 1:100,000 LA, 1:10 LA, 1:1000 LA, 1:100,000 Note: “L” designates cultures grown in LB and “LA” designates cultures grown with LB+amp. 2. Add 0.9 mL of LB broth to the tubes labeled “L, 1:10”, and “LA, 1:10.” 3. Add 1.0 mL of LB broth to the remaining four tubes. 4. Dilute the culture labeled “LB” by 1:10 as follows: agitate the tube labeled “LB” and immediately transfer 100 μL of the culture from this tube to the 0.9 mL of LB broth in the tube labeled “L, 1:10.” Mix well. Resuspending the bacteria just prior to pipetting is critical for the accuracy of the assay. 5. Transfer 10 μL of the dilute culture you just made (in the tube labeled L, 1:10) to the tube labeled “L, 1:1000.” Mix well. Again, remember to resuspend the bacteria just before pipetting. 6. Transfer 10 μL of the dilute culture in the “L, 1:1000” tube to the tube labeled “L, 1:100,000.” Mix well. As in the steps above, agitate the tube the transfer is made from just prior to making the transfer. ©2018 Carolina Biological Supply Company Fill-in • S-10 Evolution in Real Time: Bacteria and Antibiotic Resistance Laboratory Investigation (continued) STUDENT GUIDE Name Date 8. Allow the liquid to soak into the plates for a few minutes, and then flip the plates over to incubate them, lidside down, overnight at the temperature indicated by your instructor. 9. Using the same procedure as in steps 5–8, plate 20 μL of culture from the “LA, 1:100,000” tube onto the LB and LB/amp plates that you labeled “LA.” 10. Wipe down your area with 70% ethanol and wash your hands. Predict the Result Predict what type of colonies you expect to see on each plate and explain the reasoning behind each prediction. ©2018 Carolina Biological Supply Company Fill-in • S-12 Evolution in Real Time: Bacteria and Antibiotic Resistance Laboratory Investigation (continued) Name STUDENT GUIDE Date Day 9: Tabulating the Results Materials For each group: LB and LB/amp plates from day 8 permanent marker, 1 or more Procedure 1. Create a data table to record your colony counts. 2. Carefully count the number of colonies on each plate and fill in the data table. To count accurately, use a permanent marker to put a dot by each colony as you count it. If the numbers are high, try to come up with a simple strategy for keeping track of colonies as you count. Analysis 1. Questions 1a–d pertain to the plates that were plated with the culture grown in LB/amp. These plates are the plates labeled “LA.” a. What is the phenotype of the colonies on the LB/amp plate with respect to ampicillin resistance? b. What is the phenotype of the colonies on the LB plates? Explain your answer. c. How does the number of colonies on the LB/amp plate compare with the number on the LB plate? What do these relative numbers suggest to you? Explain. d. Can you think of a way to verify the phenotype of the colonies on the LB plate? Explain your method. Evolution in Real Time: Bacteria and Antibiotic Resistance Fill-in • S-13 ©2018 Carolina Biological Supply Company Laboratory Investigation (continued) STUDENT GUIDE Name Date 2. Read the following paragraph and then answer the question. Mechanisms of Antibiotic Resistance Bacteria develop resistance to antibiotics through a variety of mechanisms: •• The target of the antibiotic is modified so that the antibiotic can no longer work. For example, some antibiotics kill bacteria by binding to their ribosomes and inhibiting translation. Some bacteria are resistant to these antibiotics because their ribosomes have been altered such that the antibiotic no longer binds. •• The bacteria evolve in a way that prevents the antibiotic from entering or from staying in the cell, for example, by eliminating the surface molecules through which the antibiotics enter or by developing surface proteins that pump the antibiotic out of the cell before it reaches its target. •• Some develop ways to alter the antibiotic molecule itself so that it no longer binds to its target efficiently. For example, if an antibiotic binds to the bacterial ribosome to stop bacterial growth, the bacteria will alter the antibiotic, so it can no longer bind. •• Some produce enzymes that are able to destroy the antibiotic. For example the beta-lactamase made by the ampicillin-resistance bacteria used in this kit destroys ampicillin. The bacteria also secrete the beta-lactamase into the surrounding medium. How did the observed phenotypic ratios for the bacterial culture grown in the presence of ampicillin come about? Give a step-by-step explanation. Include any of the information in the paragraph above that is relevant to your argument. 3. Questions 3a–e pertain to the plates plated with the culture grown in LB. The plates are labeled “L.” a. With respect to ampicillin resistance, what is the phenotype of the colonies on the LB/amp plate? Explain the results that you got. b. What is the likely phenotype of the colonies on the LB plates? Explain your answer. ©2018 Carolina Biological Supply Company Fill-in • S-14 Evolution in Real Time: Bacteria and Antibiotic Resistance Laboratory Investigation (continued) Name STUDENT GUIDE Date c. Compare the number of colonies on the LB/amp plate and the LB plate. d. What does that comparison suggest about how the bacterial population in the culture evolved when grown in the environment without ampicillin? e. Why do you think the ampicillin-sensitive bacteria came to dominate the culture? Put another way, in this environment what advantage might ampicillin-sensitive bacteria have over ampicillin-resistant ones? Read back through the introductory material for a clue. 4. A student performs the experiment in this lab and finds that there are still ampicillin-sensitive colonies in the culture after the culture has been grown in the presence of ampicillin for a week. Explain how this would be possible. 5. A single E. coli bacterium like the ones used in this kit can divide into two new cells in 20 minutes when grown at 37°C. a. How many generations does the bacterial population go through in 24 hours? Evolution in Real Time: Bacteria and Antibiotic Resistance Fill-in • S-15 ©2018 Carolina Biological Supply Company Laboratory Investigation (continued) STUDENT GUIDE Name Date b. How many progeny can a single bacterium produce in 24 hours? Assume that growth conditions are not limiting (i.e., the bacteria do not run out of food and space). 6. During the 7 days you grew your cultures, the bacteria go though many generations. Do you think the relative numbers of ampicillin-sensitive and ampicillin-resistant bacteria in the culture grown in the absence of antibiotic for 7 days would be different from what you observed if each bacterium required 6 days to divide? Why? For this question, assume that the bacterium cannot lose the plasmid. 7. A correlation is a relationship between two variables, but that relationship is not necessarily causative; in other words, a change in one of the variables does not necessarily cause a change in the other. For example, a rise in the number of car crashes over a period of five years might correlate almost exactly with a decrease in expenditure on highway maintenance. The correlation does not necessarily mean that the neglect caused the crashes. Further analysis might indicate that the cause is something entirely different, such as a change in speed limit or a general increase in the number of cars and drivers on the roadways. Make an argument for why the experiment you performed in this lab demonstrates that the loss of antibiotic resistance by the bacteria is truly caused by the absence of the ampicillin in the medium. ©2018 Carolina Biological Supply Company Fill-in • S-16 Evolution in Real Time: Bacteria and Antibiotic Resistance Assessment Name STUDENT GUIDE Date 1. Given what you observed in this lab, would you argue for or against treating livestock with an antibiotic even when they show no sign of disease? Explain your reasoning. 2. Reread the description of the phenomenon at the beginning of the lab. Given the information in that description and what you have observed in the lab, come up with a hypothesis for how plasmid-mediated colistin resistance came to be found in bacteria infecting humans. In writing your hypothesis, try to use terms that are used when discussing evolution. 3. A patient comes to the doctor’s office with symptoms identical to those of a routine respiratory infection that is in the community. The respiratory infection is caused by a virus, not a bacterium. Viruses are not killed by antibiotics. The patient wants to be given antibiotics. You are the doctor. Give two good reasons why the patient should not have antibiotics. 4. There are two different hog farms, farm “A” and farm “B.” The hogs are raised the same way on each farm, with one exception. On farm A, a small amount of antibiotic “D” is always added to their feed. This increases the hogs’ growth rate, possibly by reducing infections. On farm B, no antibiotics are included in the animals’ feed. a. What trait is likely to become more prevalent among the bacteria found in the hogs’ immediate environment on farm A? Explain why. b. On which farm is it more likely that antibiotic D will effectively treat any bacterial infection that may arise in the hogs? Why? Evolution in Real Time: Bacteria and Antibiotic Resistance Fill-in • S-17 ©2018 Carolina Biological Supply Company Assessment (continued) STUDENT GUIDE Name Date 5. To combat the problem of antibiotic resistance, researchers try to develop new antibiotics that will kill or stop the growth of bacteria in novel ways. Aside from a knowledge of chemistry (necessary if the antibiotics are synthesized or are modified from existing molecules), what kind of knowledge would be useful in developing new antibiotics? 6. You are studying two different patches of forest with almost identical environments. Each patch contains a population of a lizard species with both striped and solid-colored individuals. There are approximately equal numbers of striped and solid lizards in each forest patch. A population of birds that show a preference for eating the solid-colored lizards moves into forest patch A. The birds do not reach patch B. With respect to the striped trait, how do you expect the lizard population in the two patches of forest to evolve? Explain. ©2018 Carolina Biological Supply Company Fill-in • S-18 Evolution in Real Time: Bacteria and Antibiotic Resistance