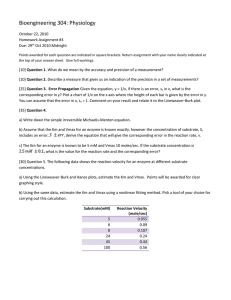
Sample Exam Questions for “A Likely Story” Enzyme Inhibition Case Study 1. The inhibitor binds to the enzyme-substrate complex or to the free enzyme equally well. This statement describes which of the following types of inhibition? (Tymoczko 3E Section 8.2, Pgs. 134-136 and Figure 8.9) a. Irreversible b. Competitive c. Noncompetitive d. Mixed 2. The graph on the right represents a Lineweaver-Burk Plot for a given enzyme. The Vmax for this enzyme is approximately: (Tymoczko 3E Section 7.2, Pg.115, Figure 7.5 and Equation 10) a. 0.25 b. 0.5 c. 1 d. 2 e. Vmax cannot be determined from this kind of plot 10 8 6 4 2 0 0 1 2 3 4 5 5 velocity 3. Calculate KM using the data shown in the plot on the right: (Tymoczko 3E Section 7.2, Pg.115, Figure 7.5 and Equation 10) a. 0.1 mM b. 0.2 mM c. 2.0 mM d. 4.0 mM e. KM cannot be determined from this data 12 4 3 2 1 0 0 0.5 1 1.5 2 [S] mM 4. 5. At [I] = 0, KM = 0.5 nM and Vmax = 0.6 mmole/s. At [I] = 5 mM, KM = 0.1 nM and Vmax = 0.2 mmole/s. What type of inhibitor is this? (Tymoczko 3E Section 8.2, Pgs. 135-136 and Figures 8.10-8.12) a. Competitive b. Noncompetitive c. Uncompetitive d. Irreversible e. Transition state analog At what substrate concentration would an enzyme with a kcat of 30.0 s-1 and a KM of 0.0050 M operate at one-quarter of its maximum rate? (Tymoczko 3E Section 7.2, Pg. 116 and Equations 13 and 14) a. [S] = 2KM b. [S] = ½ KM c. [S] = 1/3 KM d. [S] = ¼ KM e. Cannot determine with information given 6. In an enzyme assay, you determine the KM for your enzyme to be 0.5mM. You measure the maximum rate of the reaction to be 0.1 mmoles/second. Calculate kcat if the above experiment was conducted in a 1.0 mL reaction volume at an enzyme concentration of 20 nM. (Tymoczko 3E Section 7.2, Pg. 116 and Equations 13 and 14) kcat = Vmax/Etotal [E] = 20 nM Etotal = [E] x Volume Etotal = (20 nM)(1mL) Etotal = 2 x 10-11 moles Vmax = 0.1 mmoles/second kcat = Vmax/Etotal kcat = (0.1 mmoles/second) / (2 x 10-11 moles) kcat = 5,000,000 s-1 7. You discover a new inhibitor to an enzyme and a crystal structure clearly shows that it binds in the active site at the same location as the substrate. What do you think that this means about the likelihood that this inhibitor will show competitive inhibition? How would you know if this inhibitor was competitive or not? (Tymoczko 3E Section 8.2, Pgs. 135-136 and Figure 8.10) The inhibitor is almost certainly a competitive inhibitor. Kinetics data showing that the inhibitor decreases KM while leaving Vmax the same would confirm this conclusion. 8. . Congratulations! You have just been hired as a biochemist for the company Hinesbiopharma! You have been assigned to work on an exciting project studying an enzyme known as ABCase. Others have determined the KM for the enzyme to be 0.50 mM. Your first task is to determine kcat. Adding 5.0 μL of a 2.0 mg/mL enzyme stock solution to an assay mixture with a 1.0 mL total volume, you measure a Vmax of 0.60 umol/min. Taking into account that ABCase is a monomeric enzyme with a molecular mass of 45,000 daltons, what is the kcat of ABCase? (Tymoczko 3E Section 7.2, Pg. 116 and Equations 13 and 14) 45 s-1 9. The graph shown to the right represents an enzyme assay (note the units! P stands for “product”); use it to answer the following questions: (Tymoczko 3E Section 7.2, Pg. 113114 and Figure 7.3, Equations 5,6, & 7) a. Can Vmax and KM be estimated using this data? If so, what are they? If not, why not? Cannot. Graph is a reaction progress curve, not a Michaelis plot. b. Assuming that initially there was 0.3 mM substrate present in the assay, what is going on in this system around the time indicated by the blue arrow? Substrate is not completely consumed. System has reached dynamic equilibrium. 10. Hinesbiopharma has created a compound called CH523 that binds to an allosteric site approximately 15Å away from the active site of ABCase. Crystal structures reveal that this allosteric site is blocked in the absence of substrate; a Phe residue blocks the ligand binding pocket. When the enzyme binds the substrate, a shift in a helix connecting the active site and the allosteric site displaces the Phe sidechain, clearing the pocket and allowing the inhibitor to bind. When the inhibitor is bound, it prevents a nearby loop from moving to associate with the remainder of the active site. The association of this loop with the active site is necessary for catalysis to occur. Binding studies have confirmed predictions made by the crystal structures: in the absence of substrate, the inhibitor does not bind to the enzyme. What type of kinetics do you think CH523 will exhibit? Explain your answer! (Tymoczko 3E Section 8.2, Pgs. 134-136 and Figures 8.10-8.12) Uncompetitive