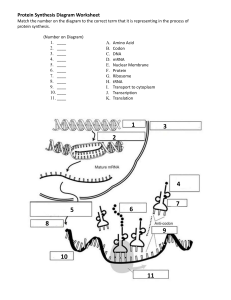
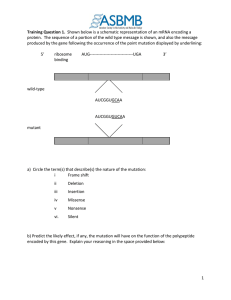
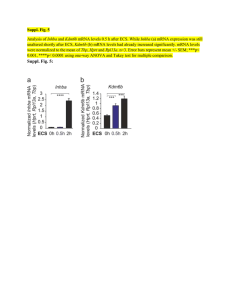
CHAPTER 40 The Mechanism of Protein Synthesis Problems: 2,3,6,7,9,13,14,15,18,19,20 • Initiation: Locating the start codon. • Elongation: Reading the codons (5’→3’) and synthesizing protein amino→carboxyl. • Termination: Recognizing terminal codon and releasing protein. Prokaryotes and Eukaryotes 40:1 Decoding the Information in mRNA Exit Peptidyl Aminoacyl Initiation Start codon (also GUG, Val) Shine-Delgarno Sequence Start: 1. Pairing of 16S rRNA with SD of mRNA (a UTR). 2. Pairing of initiator tRNA with start codon. Initiator tRNAf Two different tRNA’s that carry Met; tRNAf and tRNAm. One Synthetase adds Met to both tRNAf and tRNAm. A specific transformylase only formylates Met-tRNAf. Initiation Initiation factors, IF1, IF2 and IF3. IF1 and IF3 prevent premature assembly of the 70S ribosome. IF2 binds to fMet-tRNAf and mRNA at AUG codon. The fMet-tRNAf in the fully assembled ribosome is in the P-site. Initiation Elongation Factor, EF-Tu brings appropriate aa-tRNA to the A-site. fMet AUG Requires GTP: EF-Tu-GTP Protects aa-tRNA ester bond. GTP→GDP only when proper codon is present and then EF-Tu is released. 40:2 Peptidyl Tranferase and Peptide Bond Formation---Elongation Peptidyl tranferase center on 23S rRNA of 50S subunit. Catalysis occurs via mechanism of proximity and orientation. EF-Tu-GTP EF-Ts GDP GTP -EF-Tu-GTP -EF-Tu-GDP EF-Tu-GDP Peptide grows from the H3N+-end to –COOH-end Termination Release Factors (RF) recognize the stop codons: RF1-UAA or UAG and RF2-UAA or UGA. RF recognizes stop codon and interacts with peptidyl transferase promoting addition of H2O, rather than aminoacyl-tRNA, to the growing peptide chain. GTP hydrolysis after binding EF-G and binding of ribosome release factor (RRF) cause release of ribosome. 40:3 Bacteria and Eukaryotes Differ in Initiation of Protein Synthesis Larger ribosome: 80S versus 70S. Initiator tRNAi and Met-tRNAi is not formylated; differs from Met-tRNAm. Initiation involves eIF’s, CAP recognition (eIF-4E), and movement to AUG start codon (eIF-2). Circular structure of eukaryotic mRNA thought to facilitate rebinding of ribosomes Important control point. Mutation leads to serious disease. Elongation: EF1 and EF1 are counterparts of EF-Tu and EF-Ts, and EF2 is equivalent of bacterial EF-G translocase. Termination: eRF1 is the release factor and is the only release factor in eukaryotes. eIF-3 prevents premature ribosome reassembly in absence of initiation factors. Organization: In eukaryotes the translation machinery is organized as a large complex with the cytoskeleton. 40.4: Antibiotic Protein Synthesis inhibitors. Diphtheria toxin inhibits EF2 (the eukaryotic translocase). 40:5 Secretory and Membrane Proteins Protein targeting or sorting: 1. Posttranslational delivery to nucleus, chloroplast, mitochondria, and peroxisomes. 2. Secretory pathway to ER, and then the Golgi, lysosomes, integral membrane proteins, etc. Signal recognition particle Packaged in transport vesicles and transported to location via exo- and endocytosis 40:6 Protein Synthesis Regulation Regulation by use of mRNA: Iron Response Elements (IRE) Ferritin mRNA contains an IRE to which an IRE binding protein (IRE-BP) binds and blocks translation of the mRNA. High iron: IRE-BP binds iron and can not bind ferritin mRNA. Ferritin is produced to store iron. Low iron: IRE-BP binds ferritin mRNA and blocks ferritin production Transferrin-receptor mRNA contains an IRE to which an IRE binding protein (IRE-BP) binds under low iron that stabilizes the mRNA, permitting translation and production of the receptor. Regulation Through Small RNAs RNA interference, RNAi: External source of dsRNA is cleaved into 21-nucleotide fragments by an RNase → ss siRNA → RISC which locats siRNA onto complementary mRNA and promotes their degradation. MicroRNAs, miRNA: miRNA produced from endogenous encoded RNA precursor this binds to complementary region of mRNA forming a 21-nucleotide ds RNA region which promotes degradation of that mRNA. Human genome has more that 700 miRNAs, 60% of genes regulated by one or more miRNAs: cell differentiation, development, disease such as cancer, etc.