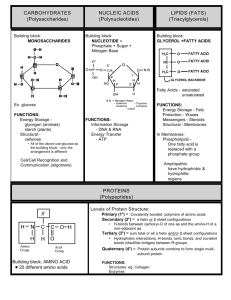
THERMODYNAMICS Thermodynamics is the study of (energetics of chemical reactions). There are 2 relevant forms of energy in chemistry: (kinetic energy) which looks at (the movement of molecules) and (potential energy) which looks at (energy stored in molecules). (ATP) is the most important energy storage molecule in all cells. It stores energy in its (ester bonds between phosphate groups). The first law of thermodynamics, also known as the law of conservation of energy, states that (the energy of the universe is constant, so if the energy of a system decreases, the energy of the surroundings must increase and vice versa). The second law of thermodynamics states that (the disorder or entropy of the universe tends to increase). A practical way to discuss free energy is by looking at Gibbs free energy. The formula for Gibbs free energy is (∆𝐺 = ∆H − T∆S) where S stands for (entropy), T stands for (temperature), and H stands for (enthalpy) which can be defined by another equation (∆𝐻 = ∆𝐸 + 𝑃∆𝑉) where E stands for (bond energy of products and reactants in a system), P stands for (pressure) and V stands for (volume). ∆G (increases) with increasing ∆H and (decreases) with increasing entropy. The change in Gibbs free energy determines if a reaction is (spontaneous or nonspontaneous). If ∆G is positive, the reaction is (nonspontaneous) if ∆G is negative, the reaction is (spontaneous). In other words, Gibbs free energy shows whether or not the reactants will stay as they are or be converted to products. Reactions with a -∆G are spontaneous and (exergonic) meaning (energy exits the system). Reactions with ∆G are nonspontaneous and (endergonic) meaning (energy has to be added). Reactions with a -∆H are called (exothermic) which means they (release) heat. Reactions with a ∆H are called (endothermic) which means they (absorb) heat. When all reactants and products are present at a 1M concentration, chemist calculate a standard free energy change denoted (∆𝐺°). Under physiological conditions, chemist use (∆𝐺°′). This can be related to the equilibrium constant for a reaction by using the following equation, ! (∆𝐺°! = −𝑅𝑇𝑙𝑛𝐾"# ). K’eq is the (ratio of products to reactants at [%]!" [']!" equilibrium) and can be expressed ( [(]!" [)]!" ) To calculate ∆𝐺 for a reaction in the body, we would use the following equation, (∆𝐺 = [%]['] ∆𝐺°! + 𝑅𝑇𝑙𝑛𝑄) where (𝑄 = [(][)]), using the actual concentrations of A,B, C, and D. The difference between Q and Keq is that Q is (the ratio of products to reactants at any given set up), while Keq is (the ratio of products to reactants at equilibrium). All reactions will eventually reach equilibrium however, if the reaction is disturbed by adding more product/reactant or taking away product/reactant, there will be a change in Q and not Keq. Ultimately, the reaction will proceed in the direction to reestablish equilibrium. Equilibrium is (the point at which concentrations stay the same, the rate if the reaction in the forward direction = the rate of the reaction in the reverse direction). In summary, there are two factors that determine whether a reaction will occur spontaneously within a cell. 1. (Its equilibrium constant Keq) 2. The concentrations of reactants and products RTlnQ In a lab there is a third factor which is (temperature). NOTE: Spontaneous means that a reaction is (energetically favorable) but says nothing about the (rate of the reaction). ∆𝐺 only depends on the difference of energies, not the pathway. KINETICS AND ACTIVATION ENERGY The reason some spontaneous reactions proceed very slowly or not at all is due to (the large amount of energy required to get them going). The study of reaction rates is called chemical kinetics. All reactions proceed through a transition state that is (unstable and takes a great deal of energy to produce). The energy required to produce a transition state is called (activation energy). A lower activation energy makes for (a faster rate of reaction) while a higher activation energy makes for (a slower rate of reaction). A (catalyst) lowers the Ea of a reaction without changing the ∆𝐺. It does this by (stabilizing the transition state) making it less thermodynamically unfavorable). The second important characteristic of a catalyst is that it is (regenerated) with each reaction cycle. (Enzymes) are biological catalysts. OXIDATION AND REDUCTION Plants and animals store chemical energy in reduced molecules such as carbohydrates and fats. These reduced molecules are (oxidized) to produce (CO2 and ATP). The energy of (ATP) is used in turn to (drive the energetically unfavorable reactions of the cell). (Oxidation) is a chemical term meaning (the loss of electrons). Reduction means (the gain of electrons). Molecules can gain or lose electrons depending on the other atoms they are bound to. Recognizing oxidation reactions: 1) (gain of oxygen atoms) 2) (loss of hydrogen atoms) 3) (loss of electrons) Recognizing reduction reactions: 1) (loss of oxygen atoms) 2) (gain of hydrogen atoms) 3) (gain of electrons) NOTE: When one atoms gets reduced, another must be (oxidized). This is called a (redox pair). (Catabolism) is the process of breaking down molecules. (Anabolism) is the process of building up molecules. For example, when we extract energy from glucose, we are doing so via (oxidative catabolism). We break down the glucose by oxidizing it. As we oxidize food, we release the stored energy plants got from the sun. We store this energy in the form of (ATP). We can use the energy in ATP to generate storage molecules such as glycogen and fatty acids. Fatty acids are generated by successive reduction of a carbon chain. Thus, (anabolic) processes are generally reductive. ACIDS AND BASES DEFINITIONS: Bronsted Lowry acids are (proton ditchers) while Bronsted Lowry bases are (proton acceptors). Lewis acids are (electron pair acceptors) while Lewis bases are (electron pair donors). When a Bronsted Lowry acids donates an H+, its remaining structure is called a (conjugate base). Likewise, when a Bronsted Lowry base gains a H+, its new structure is called a (conjugate acid). STRENGTHS OF ACIDS/BASES: The strength of an acid is directly related to how much the products are favored over the reactants. This makes sense, because stronger acids are more likely to dissociate! A reaction between a generic acid and water is written as (𝐻𝐴(𝑎𝑞) + 𝐻* 𝑂(𝑙 ) ↔ 𝐻+ 𝑂, (𝑎𝑞) + 𝐴- (𝑎𝑞)). The equilibrium expression for an acid dissociating in water reaction is (𝐾. = [/# 0$ ][(% ] [/(] ). Ka is known as the (acid-ionization or acid-dissociation constant) of the acid HA. We can rank acid strength by comparing their Ka values. The larger the Ka value, (the stronger the acid) and the smaller the Ka value, (the weaker the acid). We can apply these same ideas to the strength of bases. A reaction between a generic base and water is written as (𝐵(𝑎𝑞) + 𝐻* 𝑂(𝑙 ) ↔ 𝐻𝐵, (𝑎𝑞) + 𝑂𝐻- (𝑎𝑞)). The equilibrium expression for a base dissociating in water reaction is (𝐾1 = [/)$ ][0/ % ] [)] ). Kb is known as the (base-ionization or base-dissociation constant) of the base B. We can rank base strength by comparing their Kb values. The larger the Kb value, (the stronger the base) and the smaller the Kb value, (the weaker the base). NOTE: substances that can act as either an acid or a base are called (amphoteric). pH: The pH scale measures (the concentration of H+ (or H3O+) ions in a solution. The formula for pH given this information is (𝑝𝐻 = −log [𝐻, ]). A basic solution will have a pH (greater than 7), a neutral solution will have a pH (of about 7), and an acidic solution will have a pH (less than 7). A low pH indicates a (high) concentration of H+ while a high pH indicates a (low) concentration of H+. Alternatively, acidity/basicity can be measured in terms of hydroxide ion concentration [OH-] by using (pOH). The formula for pOH is (𝑝𝑂𝐻 = −log [𝑂𝐻- ]). A high pOH means the solution is (acidic) while a low pOH means the solution is (basic). To determine pH from pOH and vis versa, we can use the following formula (pH + pOH = 14). RELATIONSHIPS BETWEEN CONJUGATES: In general, “p” of something is equal to (the -log) of that something. Therefore, pKa = (-log Ka) and pKb = (-log Kb). The lower the pKa value, the (stronger) the acid and the higher the pKa value, the (weaker) the acid. Same goes with bases. The lower the pKb value, the (stronger) the base and the higher the pKb value, the (weaker) the base. BUFFER SOLUTIONS: A buffer is a solution that (resists changing pH when a small amount of acid or base is added. The buffering capacity comes from the presence of (a weak acid and its conjugate base or a weak base and its conjugate acid) in roughly equal concentrations. The most important buffer system in our blood plasma is the (bicarbonate buffer system) consisting of (carbonic acid) and its conjugate base (bicarbonate). Carbonic acid is formed though (our cells producing carbon dioxide as a by product, and that CO2 reacting with water in a reversible fashion to form carbonic acid. AMINO ACIDS (Proteins) are biological macromolecules that act as enzymes, hormones, receptors, channels, transporters, antibodies, and support structures inside and outside cells. Proteins are composed of 20 different (amino acids) linked together in polymers. The (composition and sequence) of amino acids in the poly peptide chain is what makes each protein unique. STRUCTURE AND NOMENCLATURE: The general structure of an amino acid includes (an amino group, a carboxyl group, a hydrogen, and a variable R group). Amino acids can be organized into 4 broad categories including (acidic, basic, polar, and nonpolar). ACIDIC AMINO ACIDS: the amino acids (aspartic acid, and glutamic acid) have acidic (carboxylic acid) within their side chains. Their deprotonated/anionic forms are called aspartate and glutamate respectively. BASIC AMINO ACIDS: the amino acids (lysine, arginine, and histidine) have basic R-group side chains. At physiological pH, Lys and Arg are (cationic or protonated). His on the other hand can be protonated or deprotonated making it a readily available (proton donor and acceptor or acid/base). NONPOLAR AMINO ACIDS: the amino acids (glycine, alanine, valine, leucine, isoleucine, phenylalanine, and tryptophan) are all nonpolar and thus (hydrophobic). Sulfur-containing (methionine) is also nonpolar as well as (proline) with its unique ring structure. Nonpolar amino acids have either an (alkyl or aromatic) side chain. NOTE: (tyrosine) has an aromatic side chain but is polar. Because these amino acids don’t associate with water, you are likely to find them on the (interior of proteins away from water). The larger the hydrophobic group, the greater it repels water. POLAR AMINO ACIDS: Polar amino acids are characterized by having an R group polar enough to form (hydrogen bonds) with water but does not act like an acid or base. This means they are (hydrophilic) and will interact with water when possible. This group includes (serine, threonine, tyrosine, asparagine, glutamine) and one sulfur-containing amino acid (cysteine). AMINO ACID REACTIVITY: amino acids are amphoteric meaning (they can act as acids or bases). This makes sense as w/in the amino acid backbone, there is an acidic (carboxylic acid group) and a basic (amino group). Carboxyl groups of amino acids generally have a pKa of about (2) which is makes it a (stronger acid) relative to the ammonium groups which generally have a pKa of about (9-10). The mathematical formula that describes the relationship between pH, pKa, and the position of equilibrium in an acid-base reaction is known as (the HendersonHasselbalch equation) and is written as follows: (𝑝𝐻 = 𝑝𝐾. + log [(% ] [/(] = 𝑝𝐾. + log [1.2" 4567] [.89: 4567] ) Given the pH and pKa, we can calculate (the ratio of the base and acid forms of a compound in equilibrium) - When the pH of a solution is less than the pKa of an acidic group, the acidic group, than the acidic group will most likely be in its (protonated) form - When the pH of a solution is greater than the pKa of an acidic group, the acidic group, than the acidic group will most likely be in its (deprotonated) form APPLICATION OF ACID/BASE CHEMISTRY TO AMINO ACIDS: In its protonated or acidic form, the amine group of an amino acid is called (an ammonium group) and has a pKa of (between 9-10). THE ISOELECTRIC POINT OF AMINO ACIDS: There is a pH for every amino acid at which it has no overall net charge, aka positive and negative charges cancel. A molecule with positive and negative charges that balance is referred to as (a dipolar ion or zwitterion). The pH at which the molecule is uncharged or “(zwitterionic)” is referred to as its isoelectric point (pI). Zwitter is german for hybrid implying that an amino acid at its pI has both positive and negative charges. To calculate the pI of an amino acid, aka to find out the pH value at which the + and – charges balance, for a molecule with two functional groups, (just average the pKa’s of the two functional groups). PROTEIN STRUCTURE There are two common types of covalent bonds between amino acids in proteins: The (peptide bonds) that link amino acids together into polypeptide chains and (disulfide bridges) between cysteine Rgroups. THE PEPTIDE BOND: Polypeptides are formed by linking amino acids together in (peptide bonds). A peptide bond is formed between (the carboxyl group of one amino acid and the amino group of another) with the loss of (water). An individual amino acid is termed a (residue) when it is part of a polypeptide chain. In a polypeptide chain, the N-C-C-N-C-C pattern is formed and known as the (backbone). Hydrolysis of a protein by another protein is called (proteolysis or proteolytic cleavage) and the protein that does the cutting is known as a (proteolytic enzyme or protease). THE DISULFIDE BOND: Cysteine is an amino acid with a reactive thiol in its side chain. The thiol of one cysteine can react with the thiol of another cysteine to produce a covalent sulfur-sulfur bond known as a (disulfide bond). The cysteines forming a disulfide bond may be located in the same or different polypeptide chain(s). The disulfide bond plays an important role in (stabilizing the tertiary protein structure). Once a cysteine residue becomes disulfide-bonded to another cysteine reside, it is called (cystine) instead of cysteine. PROTEIN STRUCTURE IN THREE DIMENSIONS Each protein folds into a unique three-dimensional structure that is required for that protein to function properly. Improperly folded, or (denatured) proteins are non-functional. There are four levels of protein folding that contribute to their final 3D shape. Each level of structure is dependent upon a particular type of bond. NOTE: Denaturation means to disrupt the proteins shape without breaking peptide bonds. This can be done by (extremes of pH, temperature, urea, or tonicity). PRIMARY STRUCTURE-THE AMINO ACID SEQUENCE: The simplest level of protein structure is the order of amino acids bonded to each other in the (polypeptide chain). This linear ordering of amino acid residues is known as primary structure. The bond which determines 1o structure is (the peptide bond) as it links one amino acid to the next. SECONDARY STRUCTURE-HYDROGEN BONDS BETWEEN BACKBONE GROUPS: Secondary structure refers to (the initial folding of a polypeptide chain into shapes stabilized by hydrogen bonds between backbone NH and CO groups). The two most common shapes are (ahelix and B-pleated sheets). NOTE: The unique side chain of proline causes 2 problems in polypeptide chains 1) (formation of a peptide bond with proline eliminates the only hydrogen atom on the nitrogen atom of proline) 2) (its unique structure causes it to kink the polypeptide chain) The a-helix is a favorable structure for a (hydrophobic transmembrane region) because (polar groups are bonded to each other inside the helix meaning less interaction with water). There are two types of beta sheets, ones where the adjacent polypeptide strand runs in the same direction known as (parallel) and ones where the adjacent polypeptide strand runs in opposite direction known as (antiparallel). TERTIARY STRUCTURE: HYDROPHOBIC/HYDROPHILLIC INTERACTIONS: Tertiary structure interactions involves interactions between amino acid residues located more distantly from each other within the polypeptide chain. These interactions include (van der Waals forces between nonpolar side chains, hydrogen bonds between polar side chains, disulfide bonds between cysteine residues, and electrostatic interactions between acidic and basic side chains). The folding of secondary structures into tertiary structures is driven by (interactions of R groups with each other and with the solvent-water). Hydrophobic R groups tend to fold into the (interior) of the protein, while hydrophilic R groups tend to be (exposed to water on the surface of the protein). This is called the (hydrophobic effect). QUARTERNARY STRUCTURE-VARIOUS BONDS BETWEEN SEPARATE CHAINS: 4o structure describes (interactions between polypeptide subunits). A subunit is (a single polypeptide chain that is part of a large complex containing many subunits (a multisubunit complex). The forces stabilizing quaternary structure are (van der waal forces, hydrogen bonds, disulfide bonds, and electrostatic interactions). PROTEINS AS ENZYMES: (Enzymes) are biological catalysts. They increase the rate of a reaction by (lowering the reactions activation energy) but they do NOT affect (∆𝐺) between reactants and products. Enzymes have a kinetic role not a thermodynamic role. Enzymes can be divided into different classes each involved in a type of reaction. Hydrolase (hydrolyzes chemical bonds). Isomerase (rearranges bonds within a molecule to form an isomer. Ligase (forms a chemical bond). Lyase (breaks chemical bonds by means other than oxidation or hydrolysis). Kinase (transfers a phosphate group to a molecule from a high energy carrier such as ATP). Oxidoreductase (runs redox reactions). Polymerase (runs polymerization reactions). Phosphatase (removed a phosphate group from molecule). Phosphorylase (transfers a phosphate group to a molecule from inorganic phosphate. Protease (hydrolyzes peptide bonds). ATP AS AN ENERGY SOURCE-REACTION COUPLING: Thermodynamically unfavorable reactions in the cell can be driven forward by (reaction coupling). In reaction coupling, (one very favorable reaction is used to drive an unfavorable one). This is possible because (free energy changes are additive). The favorable reaction in the cell is (ATP hydrolysis). How does ATP hydrolysis drive unfavorable reactions? 1) (by causing a conformational change in a protein) 2) (by transfer of a phosphate group from ATP to a substrate) ENZYME STRUCTURE AND FUNCTION: The reason for proper folding in enzymes, is (the proper formation of the active site). The active site is (the region of the enzyme that is directly involved in catalysis). The reactants in an enzyme-catalyzed reaction are called (substrates). What is the role of the active site/ how do enzymes work? The active site model, states (the substrate and active site are perfectly complementary). This differs from the induced fit model which states (the substrate induced a conformational change in the enzyme). Recognition pockets are pockets in the enzyme structure that (attract certain residues on substrate polypeptides). REGULATION OF ENZYME ACTIVITY: Metabolic pathways are not all continually on, rather they are regulated. Therefore, enzymes are regulated on one or more of the following ways: 1) Covalent modification: (include the addition or removal of chemical groups) 2) Proteolytic cleavage: (process of breaking the peptide bonds between amino acids in proteins) 3) Association with other polypeptides: (self-explanatory) 4) Allosteric regulation: molecule alters conformation of enzyme ALLOSTERIC REGULATION: Allosteric regulation is (the binding of small molecules to particular sites on an enzyme that are distinct from the active site. When bound the allosteric regulator cab change the conformation of the enzyme to increase or decrease catalysis). This binding is generally noncovalent and reversible. FEEDBACK INHIBITION: Negative feedback/feedback inhibition (is a type of regulation in biological systems in which the end product of a process in turn reduces the stimulus of that same reaction). Positive feedback/feedback stimulation (is when the product of a reaction leads to an increase in that reaction). ENZYME KINETICS: Enzyme kinetics is (the study of the rate of formation of products from substrates in the presence of an enzyme). The reaction rate, V-for velocity, is the amount of product formed per unit of time in mol/s. It depends on (the concentration of the substrate [S] and enzyme). The point at which an enzyme is said to be saturated is known as Vmax. When a reaction reaches this, more substrate (does not increase the reaction rate at all). The Michaelis constant Km is (the substrate concentration at which the reaction velocity is half its maximum). To find Km on the enzyme kinetics graph, (mark the Vmax on the y-axis, then divide this distance in half to find Vmax/2. Km is found by drawing a horizontal line from this point to the curve and then straight down). Km is unique for each substrate pair and give information on (the affinity for the enzyme for its substrate). If the enzyme-substrate pair has a low Km, (it means not very much substrate is required to get the reaction rate to half the maximum therefore the affinity is high). COOPERATIVITY: Cooperativity describes (the changes that occur when a binding site of one of these structures is activated or deactivated affecting the other binding sites in the same molecule. It can also be described as the increasing or decreasing affinity for binding of the other sites affected by the original binding site). Negative cooperativity, is (the decrease in binding affinity once one of the sites is bound). A (sigmoidal) curve results from positive cooperative binding. The graph shows (a rapid increase in speed. This reflects how the binding on one subunit increases the chance that the other subunits will bind to a substrate). INHIBITION OF ENZYME ACTIVITY: Enzyme inhibitors can reduce enzyme activity by a few different mechanisms including (competitive inhibition, noncompetitive inhibition, uncompetitive inhibition and mixed type inhibition). Competitive inhibitors (are molecules that compete with substrate for binding at the active site). Their inhibition can be overcome by (adding more substrate) and doesn’t diminish Vmax. Noncompetitive inhibitors (bind to allosteric sites causing a conformational change). This type of inhibition does diminish Vmax because it reduces the number of viable enzymes/active sites. Uncompetitive inhibitors (bind to the enzyme allosteric site, altering the shape of the enzyme so that even if the substrate can bind, the active site functions less effectively only binds to enzyme substrate complex). Mixed type inhibition occurs when an inhibitor can bind to either the unoccupied enzyme or the enzyme substrate complex. LINEWEAVER-BURK PLOT: The Lineweaver-Burk Plot is (a graphical representation of enzyme kinetics using the Lineweaver-Burk ; =& < <&'( equation). The Lineweaver-Burk equation is = F where the slope of the graph is ( ( ; <&'( =& <&'( ; G F[>]G + ; <&'( ), the y intercept of the graph is ), and x intercept of the graph is (− ; =& ). Recall that increasing the substrate concentration [S] (increases) the reaction rate V up to a point. With a Lineweaver-Burk plot, an increase in substrate concentration means a decrease in the inverse of the reaction rate. Can help us find an inhibitor type. CARBOHYDRATES Carbohydrates can be broken down to (CO2) in a process called (oxidation) aka (burning or combustion). This process releases a large amount of energy which is why carbohydrates serve as the principle energy source for cellular metabolism. MONOSACCHARIDES AND DISACCHARIDES: a single carbohydrate molecule is called a (monosaccharide). Two monosaccharides bonded together form a (disaccharide), a few form an (oligosaccharide) and many form a (polysaccharide). The bond between two sugar molecules is called a (glycosidic linkage). This is a covalent bond formed in a (dehydration reaction) that requires enzymatic catalysis. Polymers made from these disaccharides form important biological macromolecules. Glycogen (serves as an energy storage carbohydrate in animals), starch (is similar to glycogen and serves the same purpose in plants), cellulose (makes up wood and cotton). NOTE: Humans cannot digest cellulose because (of the large numbers of β(1,4) glycosidic linkages. These cannot be cleaved by human amylases and pass though the GI tract undigested.) LIPIDS Lipids are oily or fatty substances that play 3 physiological roles: 1) (In adipose cells, triglycerides (fats) store energy) 2) (In cellular membranes, phospholipids create a barrier) 3) (Cholesterol is a special lipid that serves as a building block for the hydrophobic steroid hormones) NOTE: Lipids are hydrophobic/lipophilic. FATTY ACID STRUCTURE: Fatty acids are composed of (long, unsubstituted, alkanes that end in a carboxylic acid). The chain is typically (14-18) carbons long and because they are synthesized two carbons at a time from acetate, only (even) numbered fatty acids are made in human cells. A fatty acid with no carbon-carbon double bonds is said to be (saturated) while fatty acids with one or more double bonds in the tail is said to be (unsaturated). These double bonds are almost always (cis). Within an aqueous solution, fatty acids are likely to interact to form (micelles). The force that drives the tails into the center is called the hydrophobic interaction. TRIACYLGLYCEROLS (TG): The storage form of fatty acid is fat. The technical name for fat is (triacylglycerol or triglyceride). A triglyceride is composed of (three fatty acids esterified to a glycerol molecule). Lipases are (enzymes that hydrolyze fats). They break down the triglycerides into (two fatty acids and a monoglyceride before absorption and ultimate reassembly into a triglyceride in the enterocyte) Fats are more efficient energy storage molecules than carbohydrates for two reasons: 1) (packing: they’re hydrophobicity allows fats to pack together much more closely than carbohydrates which carry a great amount of water) 2) (energy content: fats store much more energy than carbohydrates) LIPID BILAYER MEMBRANES: Membrane lipids are phospholipids derived from DG-P. In the bilayer, hydrophobic interactions drive its formation. Saturated fatty acid residues have (greater) van der waal interactions because (the lack of kinks make stacking easier). Unsaturated fatty acid residues increase membrane (fluidity). Decreasing length of tails (increases fluidity). Cholesterol also plays a role in membrane fluidity. At low temperatures it (increases) fluidity therefore known as membrane antifreeze. At high temperatures it (reduces) membrane fluidity. MAIN IDEA: Cholesterol keeps membrane fluidity at optimal levels. So to recap, structural determinants of membrane fluidity are: 1) (degrees of unsaturation) 2) (tail length) and 3) (amount of cholesterol) TERPRENES A terpene is a member of a broad class of compounds built from isoprene units. They may be linear or cyclic and are classified by (the number of isoprene units they contain). Examples: monoterpenes = (2), sesquiterpenes = (3), diterpenes = (4) STERIOIDS Steroids are hydrophobic and have a structure based on cholesterol. It is an important component of the lipid bilayer and is obtained from diet and synthesized in the liver. It is carried in the blood packaged with fats and proteins into lipoproteins. Steroid hormones are made from cholesterol. Two examples are testosterone and estradiol. PHOSPHOROUS CONTAINING COMPOUNDS: Phosphoric acid is an inorganic acid, aka doesn’t contain carbon, with the potential to donate 3 protons. At physiological pH, phosphoric acid is significantly dissociated existing largely in anionic form. Phosphate is also known as (orthophosphate). Two orthophosphates bound together via (an anhydride linkage) form (pyrophosphate). The P-O-P bond in pyrophosphate is an example of a (high-energy phosphate bond). This name is derived from the fact that hydrolysis of pyrophosphate is (extremely favorable). There are 3 reasons that phosphate anhydride bonds store so much energy: 1) (when phosphates are linked together, their negative charges repel each other strongly) 2) (orthophosphate has more resonance forms and this a lower free energy than linked phosphates) 3) (Orthophosphate has a more favorable interaction with water than linked phosphates) NUCLEOTIDES: Nucleotides are the building blocks of (nucleic acids) like (RNA and DNA). Each nucleotide contains a ribose/deoxyribose sugar group, a purine/pyrimidine base and 1, 2, or 3 phosphate units.