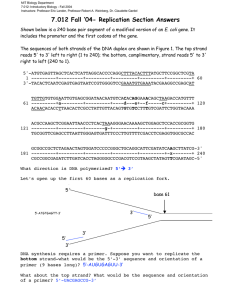
CHAPTER 3: GENE STRUCTURES AND DNA REPLICATIONS NORSHAFIQA SALIM Lesson Learning Outcome Point out gene structures and DNA replications. Distinguish the structures and modes of replications of bacterial chromosomes and plasmids. Analyze the structure of bacterial genes. Describe the replications of bacterial chromosomes (E.coli). DNA Replication Animation OVERVIEW Deoxyribonucleic 4 Acid Bases Purines Adenine Guanine Pyrimidines Cytosine Thymine* Sugar is Deoxyribose OVERVIEW Genes Genes are the basic physical and functional units of heredity. Each gene is located on a particular region of a chromosome and has a specific ordered sequence of nucleotides (the building blocks of DNA). Four requirements for DNA to be genetic material Must carry information Cracking the genetic code Must replicate DNA replication Must allow for information to change Mutation Must govern the expression of the phenotype Gene function Rosalind Franklin’s DNA image “Chargoff’s rule” A=T & C=G DNA REPLICATION Cell division and DNA replication • Cells divide Growth, Repair, Replacement • Before cells divide they have to double cell structures, organelles and their genetic information. DNA replication = Make copies of DNA Happens = When new cells need to be made DNA REPLICATION Mechanistic Overview Identical base sequences Meselson-Stahl experiments DNA replication : The 3 possible models, replication is… DNA Replication Conservative replication: One daughter helix gets both of the old (template) strands, and the other daughter helix gets both of the new (nascent) strands Semiconservative: Each daughter helix gets one old strand and one new strand Dispersive: old and new The daughter helices are mixes of Semiconservative Model Half of the parent molecule is retained by each daughter molecule. The double-stranded helix unwinds, each parent strand serves as a template for the synthesis of a complementary daughter strand. One complete strand inherited from the parental duplex, one complete strand has been newly synthesized. Replication in E. coli Replication begins at a specific site on the bacterial chromosome in a circular molecule of DNA called = origin of replication (ori) Replication proceeds outward from the origin in both directions around the bacterial chromosome = bidirectionally. Pair of replicated segments come together and join the nonreplicated segments = replication forks As the DNA helix unwinds from the origin, the two old strands become two distinctive templates: the 3´ 5´ template, and the 5´ 3´ template The replication forks eventually meet at the opposite side of the bacterial chromosome This ends replication DNA Synthesis is Bidirectional Two nascent, labeled strands at each fork means both parent strands serve as templates Bidirectional replication 1. Initiation Step DNA helicase separates the two DNA strands by breaking the hydrogen bonds between them. The replication fork moves in opposite direction, synthesize both strands simultaneously. This generates positive supercoiling ahead of each replication fork. DNA gyrase travels ahead of the helicase and alleviates these supercoils. Single-strand binding proteins bind to the separated DNA strands to keep them apart. 1. 2. 3. 4. DNA helicase Single-stranded DNA binding protein (SSB) RNA primer Primase 2. Elongation Step 2. Elongation Step Primase binds to the 1st sequence on the template & synthesize a short RNA primer. DNA polymerase III uses the primer to initiate DNA synthesis by adding deoxyribonucleotides to 3’ end. DNA polymerase join the nucleotides (phosphodiester bonds) as the new nucleotides line up opposite each parent strand by hydrogen bonds. DNA can only synthesized in 5’ to 3’ direction. 2. Elongation Step DNA polymerase can only add new nucleotides in a preexisting strand. Thus, primase capable to join RNA nucleotide without requiring preexisting strand RNA primer. In lagging strand: After a few nucleotides are added, primase replaced by DNA polymerase. DNA polymerase can now add nucleotides to 3’ end of short primer. After primer removal is complete, DNA ligase links together adjacent Okazaki fragments Leading strand vs Lagging strand: • Leading strand synthesized 5’ to 3’ in the direction of the replication fork movement. continuous requires a single RNA primer • Lagging strand synthesized 5’ to 3’ in the opposite direction. discontinuous (not continuous) requires many RNA primers , DNA is synthesized in short fragments called Okazaki fragments. 3. Termination Step Replication of bacteria terminates at opposite direction to the origin of replication (Terminus sequences). Ter sequence works as binding site for protein Tus which stops the helicase and termination of DNA replication. Then, the completed chromosomes partitioned the 2 daughter cells. The mechanism of DNA replication Arthur Kornberg et al Initiation Proteins bind to DNA and open up double helix Prepare DNA for complementary base pairing Elongation Proteins connect the correct sequences of nucleotides into a continuous new strand of DNA Termination Proteins release the replication complex Summarize Basic A. B. C. D. E. F. rules of replication Semi-conservative Starts at the ‘origin’ Can be uni or bidirectional Synthesis always in the 5-3’ direction RNA primers required Leading and Lagging strand Functions of Enzymes in Replication Helicases Primase Single strand binding proteins DNA polymerase DNA ligase - separates 2 strands - RNA primer synthesis - prevent reannealing of single strands - Add free nucleotides to synthesis new strand of DNA - seals nick via phosphodiester linkage B) Starts at origin Initiator proteins identify specific base sequences on DNA called sites of origin Prokaryotes – single origin site E.g E.coli - oriC Eukaryotes – multiple sites of origin (replicator) Prokaryotes Eukaryotes C) Uni or bidirectional Replication forks move in one or opposite directions D) Synthesis is ALWAYS in the 5’-3’ direction Nucleotide recognition Enzyme catalysed polymerisation (DNA polymerase) Complementary base pair copied Substrate used is dNTP E) RNA primers required • • DNA polymerase can only join an incoming nucleotide to one that is base-paired RNA primase provides a base paired 3’ end as a starting point for DNA pol by synthesising ~10 nucleotide primers F) Leading and Lagging Strand Anti parallel strands replicated simultaneously Leading strand synthesis continuously in 5’– 3’ Lagging strand synthesis in fragments in 5’-3’ in opposite direction. Leading and Lagging Strand New strand synthesis always in the 5’-3’ direction Animation of DNA Replication: http://highered.mcgrawhill.com/sites/0072437316/student_view0/chapter 14/animations.html http://www.learnerstv.com/animation/animation .php?ani=169&cat=biology Animation of Replication Fork: http://highered.mcgrawhill.com/sites/0072437316/student_view0/chapter 14/animations.html# http://sites.fas.harvard.edu/~biotext/animations/r eplication1.html Animation of Bidirectional Replication of DNA http://highered.mcgrawhill.com/sites/0072437316/student_view0/chapter 11/animations.html# Animation of How nucleotides are added http://highered.mcgrawhill.com/sites/0072437316/student_view0/chapter 14/animations.html#