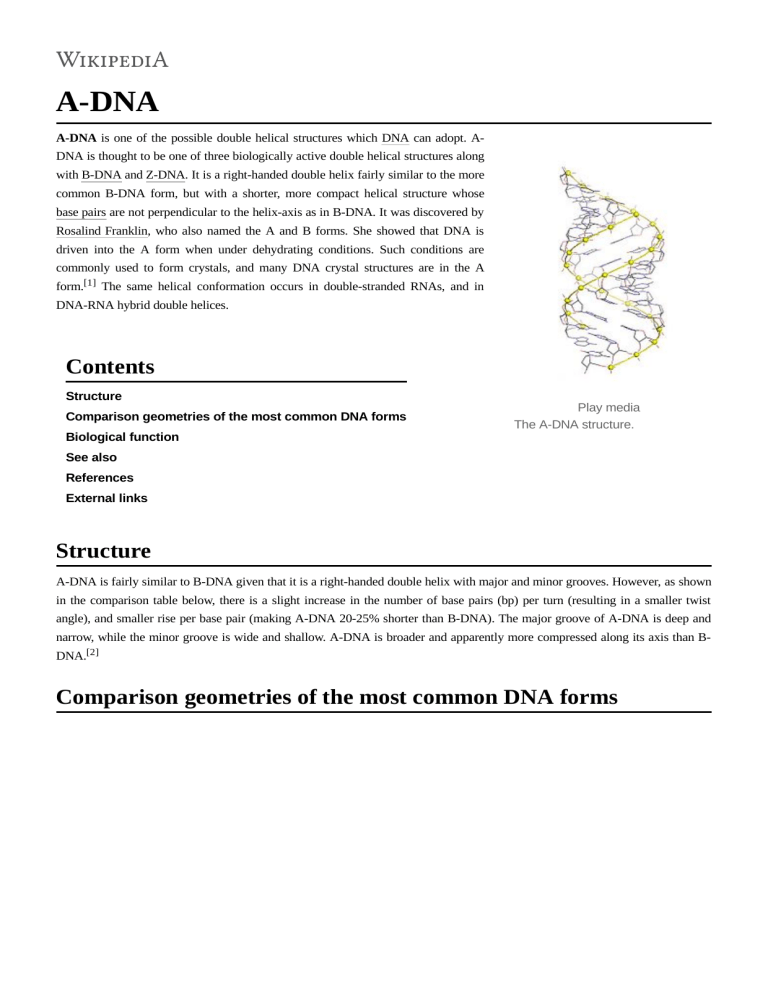
A-DNA A-DNA is one of the possible double helical structures which DNA can adopt. ADNA is thought to be one of three biologically active double helical structures along with B-DNA and Z-DNA. It is a right-handed double helix fairly similar to the more common B-DNA form, but with a shorter, more compact helical structure whose base pairs are not perpendicular to the helix-axis as in B-DNA. It was discovered by Rosalind Franklin, who also named the A and B forms. She showed that DNA is driven into the A form when under dehydrating conditions. Such conditions are commonly used to form crystals, and many DNA crystal structures are in the A form.[1] The same helical conformation occurs in double-stranded RNAs, and in DNA-RNA hybrid double helices. Contents Structure Comparison geometries of the most common DNA forms Biological function Play media The A-DNA structure. See also References External links Structure A-DNA is fairly similar to B-DNA given that it is a right-handed double helix with major and minor grooves. However, as shown in the comparison table below, there is a slight increase in the number of base pairs (bp) per turn (resulting in a smaller twist angle), and smaller rise per base pair (making A-DNA 20-25% shorter than B-DNA). The major groove of A-DNA is deep and narrow, while the minor groove is wide and shallow. A-DNA is broader and apparently more compressed along its axis than BDNA.[2] Comparison geometries of the most common DNA forms Geometry attribute: Helix sense Repeating unit A-form B-form Z-form righthanded righthanded lefthanded 1 bp 1 bp 2 bp 32.7° 34.3° 60°/2 11 10.5 12 +19° −1.2° −9° Rise/bp along axis 2.6 Å (0.26 nm) 3.4 Å (0.34 nm) 3.7 Å (0.37 nm) Rise/turn of helix 28.6 Å (2.86 nm) 35.7 Å (3.57 nm) 45.6 Å (4.56 nm) +18° +16° 0° Rotation/bp Mean bp/turn Inclination of bp to axis Mean propeller twist Glycosyl angle Nucleotide phosphate to phosphate distance Sugar pucker anti anti pyrimidine: anti, purine: syn 5.9 Å 7.0 Å C: 5.7 Å, G: 6.1 Å C2'-endo C: C2'endo, G: C3'endo C3'-endo Diameter 23 Å (2.3 nm) 20 Å (2.0 nm) Side and top view of A-, B-, and ZDNA conformations. 18 Å (1.8 nm) Biological function Yellow dots represent the location of the helical axis of A-, B-, and Z-DNA with respect to a Guanine-Cytosine base pair. Dehydration of DNA drives it into the A form, and this apparently protects DNA under conditions such as the extreme desiccation of bacteria.[3] Protein binding can also strip solvent off of DNA and convert it to the A form, as revealed by the structure of a rod-shaped virus.[4] It has been proposed that the motors that package double-stranded DNA in bacteriophages exploit the fact that A-DNA is shorter than B-DNA, and that conformational changes in the DNA itself are the source of the large forces generated by these motors.[5] Experimental evidence for A-DNA as an intermediate in viral biomotor packing comes from double dye Förster resonance energy transfer measurements showing that B-DNA is shortened by 24% in a stalled ("crunched") A-form intermediate.[6][7] In this model, ATP hydrolysis is used to drive protein conformational changes that alternatively dehydrate and rehydrate the DNA, and the DNA shortening/lengthening cycle is coupled to a protein-DNA grip/release cycle to generate the forward motion that moves DNA into the capsid. See also Mechanical properties of DNA DNA B-DNA Z-DNA C-DNA References 1. Rosalind, Franklin (1953). "The Structure of Sodium Thymonucleate Fibres. I. The Influence of Water Content" (h ttp://journals.iucr.org/q/issues/1953/08-09/00/a00979/a00979.pdf) (PDF). Acta Crystallographica. 6 (8): 673–677. doi:10.1107/s0365110x53001939 (https://doi.org/10.1107%2Fs0365110x53001939). 2. Dickerson, Richard E. (1992). DNA Structure From A to Z (https://ac.els-cdn.com/0076687992110076/1-s2.0-007 6687992110076-main.pdf?_tid=53e46970-aa00-11e7-8a59-00000aacb361&acdnat=1507230584_5ff12415fca4b 560400edc52c588d063) (PDF). Methods in Enzymology. 211. pp. 67–111. doi:10.1016/0076-6879(92)11007-6 (https://doi.org/10.1016%2F0076-6879%2892%2911007-6). ISBN 9780121821128. PMID 1406328 (https://www. ncbi.nlm.nih.gov/pubmed/1406328) – via Elsevier Science Direct. 3. Whelan DR, et al. (2014). "Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation" (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4208382). J R Soc Interface. 11 (97): 20140454. doi:10.1098/rsif.2014.0454 (https://doi.org/10.1098%2Frsif.2014.0454). PMC 4208382 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4208382). PMID 24898023 (https://www.ncbi.nlm. nih.gov/pubmed/24898023). 4. Di Maio F, Egelman EH, et al. (2015). "A virus that infects a hyperthermophile encapsidates A-form DNA" (https:// www.ncbi.nlm.nih.gov/pmc/articles/PMC5512286). Science. 348 (6237): 914–917. doi:10.1126/science.aaa4181 (https://doi.org/10.1126%2Fscience.aaa4181). PMC 5512286 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC55 12286). PMID 25999507 (https://www.ncbi.nlm.nih.gov/pubmed/25999507). 5. Harvey, SC (2015). "The scrunchworm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages" (https://www.ncbi.nlm.nih.gov/pmc/articles/ PMC4357361). Journal of Structural Biology. 189 (1): 1–8. doi:10.1016/j.jsb.2014.11.012 (https://doi.org/10.101 6%2Fj.jsb.2014.11.012). PMC 4357361 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4357361). PMID 25486612 (https://www.ncbi.nlm.nih.gov/pubmed/25486612). 6. Oram, M (2008). "Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure" (https:// www.ncbi.nlm.nih.gov/pmc/articles/PMC2528301). J Mol Biol. 381 (1): 61–72. doi:10.1016/j.jmb.2008.05.074 (htt ps://doi.org/10.1016%2Fj.jmb.2008.05.074). PMC 2528301 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2528 301). PMID 18586272 (https://www.ncbi.nlm.nih.gov/pubmed/18586272). 7. Ray, K (2010). "DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate" (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2824061). Virology. 398 (2): 224–232. doi:10.1016/j.virol.2009.11.047 (https://doi.org/10.1016%2Fj.virol.2009.11.047). PMC 2824061 (https://www.ncbi. nlm.nih.gov/pmc/articles/PMC2824061). PMID 20060554 (https://www.ncbi.nlm.nih.gov/pubmed/20060554). External links Cornell Comparison of DNA structures (https://web.archive.org/web/20061219121357/http://www.tulane.edu/~bio chem/nolan/lectures/rna/bzcomp2.htm) Nucleic Acid Nomenclature (http://jenalib.fli-leibniz.de/ImgLibDoc/nana/IMAGE_NANA.html) Retrieved from "https://en.wikipedia.org/w/index.php?title=A-DNA&oldid=883376595" This page was last edited on 15 February 2019, at 01:48 (UTC). Text is available under the Creative Commons Attribution-ShareAlike License; additional terms may apply. By using this site, you agree to the Terms of Use and Privacy Policy. Wikipedia® is a registered trademark of the Wikimedia Foundation, Inc., a non-profit organization.