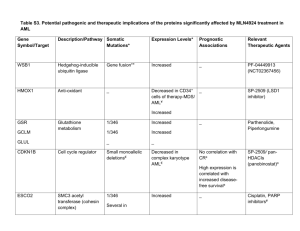
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
c-kit Expression in Human Megakaryoblastic Leukemia Cell Lines
By Zhen-Bo Hu, Weili Ma, Cord C. Uphoff, Hilmar Quentmeier, and Hans G. Drexler
A panel of 164 continuous human leukemia-lymphomacell
lines was analyzed for expression of c-kit using Northern
blotting and reverse transcriptase-polymerase chain reaction (RT-PCR). The c-kit transcripts were detectable in cell
lines assigned t o the myeloid (in 7 of 29 by Northern blotting
and in 4 of 8 by RT-PCR), monocytic (in 1 of 24 by Northern
blotting and in 3 of 6 by RT-PCR), erythroid (in 6 of 8 by
Northern blotting and in 5 of 5 by RT-PCR), and megakaryoblastic (in 10 of 10 by Northern blotting) lineages. c-kit expression was not seen by Northern blotting or RT-PCR analysis in any of the 93 lymphoid leukemia,myeloma,
or
lymphoma cell lines. Treatment of four megakaryoblastic
cell lines with protein kinase C activators (phorbol ester 120-tetradecanoylphorbol 13-acetate andBryostatin 1) led to
terminal differentiation asassessed by morphologic alter-
ations, changes in the surface marker profile, and growth
arrest. These effects were associated with enhanced c-kit
mRNA expression. Exposuret o all-trans retinoic acid downregulated c-kit mRNA levels, while simultaneously causing
morphologic alterations in all four cell lines. Stimulation
with growth factors (interleukin-3, granulocytemacrophagecolony stimulating factor, and insulin-like growth factors I
and 11). used t o assess any role of c-kit in proliferative processes, did notlead t o signifiiant upregulation or downregulation of c-kit expression. The finding of constitutive and
high expression ofc-kitmRNA in all megakaryoblastic Ieukemia cell lines and its modulation by various reagentsmight
further contribute t o the understandingof megakaryopoietic
proliferation, differentiation, and leukemogenesis.
0 1994 by The American Society of Hematology.
T
kinase C activators such as 12-0-tetradecanoylphorbol 13acetate (TPA) and Bryostatin 1 (Bryo 1) as well as alltrans retinoic acid (ATRA) to induce several cell lines to
differentiate to more mature stages. Furthermore, the hematopoietic cytokines IL-3, GM-CSF, insulin-like growth factor
I (IGF-I), and IGF-I1 were applied in attempts to probe the
effects of these molecules on c-kit gene expression during
induced proliferation of megakaryoblastic cells.
HE HUMAN PROTO-ONCOGENE c-kit encodes a
transmembrane protein known as the receptor for the
recently cloned stem cell factor (SCF) that is thought to
play a critical role in the regulation of cell proliferation and
differentiati~n.”~
Studies on mice with mutations in the W
locus showed several lesions in the c-kit gene leading to
anemia, to mast cell deficiency and reduction of progenitors
within various hematopoietic lineages, and to sterility and
lack of pigmentation; the latter aspect suggested an important
role for c-kitin hematopoiesi~?*~<~
This SCF receptor displaying tyrosine kinase activity was found to be highly expressed in various hematopoietic cells and in other tissues.2.3.6-9
Among other effects, SCF was able to stimulate
the proliferation of murine mast cells and to enhance the
formation of granulocyte-macrophage colonies, erythroid
bursts, and colonies of multipotential progenitor^.^"' The ckit ligand acted on the hematopoietic progenitor cells independently or synergistically with other factors such as interleukin-3 (IL-3), IL-6, IL-7, erythropoietin, granulocytemacrophage colony-stimulating factor (GM-CSF) and granulocyte-CSF (G-CSF).’2”5
It has been proposed that c-kit and its ligand might play a
role in the pathogenesis of acute myeloid leukemia (AML).I6
Several recent studies provided confirmatory evidence for
the hypothesis that c-kit and its ligand are involved in the
clonogenic growth of AML b 1 a ~ t s . I ~
However,
”~
clearly, less
is known about the role of c-kit and SCF in human megakaryoblastic leukemia. Biologic studies on megakaryopoiesis and
megakqocyte differentiation have been hampered
by the scarcity of megakaryocytesinthebone
m m w (BM), because
human megakaryocytes constitute less than
1% of the nucleated
cells intheBM. In contrast to this heterogeneityof cellular
components in primary material, cell lines provide the convenient advantage of a homogeneous, clonal cell population.
In the present study, we have investigated c-kit gene expression at the transcriptional level in a large panel of different continuous human leukemia-lymphoma cell lines using
Northern blot analysis and reverse transcriptase-polymerase
chain reaction (RT-PCR). We found thatall megakaryoblastic leukemia cell lines expressed c-kit mRNA, most of
them at high levels. To investigate whether expression of
the SCF receptor is associated with the differentiation of
human megakaryoblastic leukemia cells, weused protein
Blood, Vol 83, No 8 (April 15). 1994: pp 2133-2144
MATERIALS AND METHODS
Cell Culture
Cell lines were grown in appropriate media (GIBCO, Eggenstein,
Germany) supplemented with 10% to 20% heat-inactivated fetal
calf serum (FCS; Sigma, Deisenhofen, Germany) at 37°Cin an
atmosphere of 5 % COz in air. Growth factor-dependent cell lines
were cultured with the respective cytokines. The cells were examined
daily in the culture flasks under an inverted microscope. All cultures
were free of mycoplasma contamination. Cells were harvested in
their logarithmic growth phase with viabilities of higher than 90%
as examined by trypan blue exclusion.
In Vitro Treatment
The four megakaryoblastic leukemia cell lines, CMK,MDAMI?’
M-07e:’ and MKPL-l ,23 were used inexperiments aimed at modulation of c-kit gene expression by inducers and growth factors. The
cells were exposed to the following agents at 10” mom each: TPA
(Sigma), Bryo 1 (kindly provided by Prof G.R. Pettit, Arizona Cancer Center, Tempe, M ) , and ATRA (Sigma). The reagents were
From the DSM-German Collection of Microorganisms and Cell
Cultures, Department of
Human
and
Animal
Cell Cultures,
Braunschweig, Germany.
Submitted July 13, 1993; accepted December 10, 1993.
Z.B.H. was supported by the Alexander von Humboldt-Foundation.
Address reprint requests to Hans G. Drexler. MD, PhD, DSM,
German Collection of Microorganisms and Cell Cultures. Mascheroder Weg l B, D-38124 Braunschweig, Germany.
The publication costs of this article were defrayed in part by page
charge payment. This arricle must therefore be hereby marked
“advehsement” in accordance with 18 U.S.C. section 1734 solely to
indicate this fact.
0 1994 by The American Society of Hematology.
0006-4971/94/8308-02$3.00/0
2133
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
2134
HU ET AL
Table 1. Expression of c-kit mRNA in Leukemia-Lymphoma Cell Lines
Cell Line
Lymphoid leukemia cell lines
207
380
697
ALL-l
ALL-MIK
BALM-1
BALM-6
BALM-8
BAY-91
BE-13
BONNA-12
BV-173
CCRF-CEM
CML-T1
DU-528
EHEB
EH
ESKOL
EU-1
HAIR-M
HAL-01
HC-1
HK
IARC-318
JURKAT
JVM-2
JVM-3
JVM-13
KE-37
LAZ-221
LILA-1
LK-63
LOUCY
MKB-1
MN-60
MOLT-3
MOLT-l3
MOLT-l5
MOLT-l6
MOLT-l7
MT-1
NALM-l
NALM-6
OM9;22
P12/1CHIKAWA
PC-53
PEER
PF-382
REH
RS4; 11
SKW-3
SUP-B27
TAHR-87
TOM-l
Myeloma cell lines
EJM
IM-9
KARPASBZO
L-363
LP-l
Type/Origin*
Pre-B (ALL)
Pre-B (ALL)
Pre-B (ALL)
Pre-B (ALL)
Pre-B (ALL)
B (ALL)
B (ALL)
B (ALL)
Pre-B (ALL)
T (ALL)
B (HCL)
Pre-B (CML-BC)
T (ALL)
T (CML-BC)
T (ALL)
B (CLL)
B (HCL)
B (HCL)
Pre-B (ALL)
B (HCL)
Pre-B (ALL)
B (HCL)
B (HCL)
Pre-B (ALL)
T (ALL)
B (PLL)
B (PLL)
B (PLL)
T (ALL)
Pre-B (ALL)
Pre-B (ALL)
Pre-B (ALL)
T (ALL)
T (AML)
B (ALL)
T (ALL)
T (ALL)
T (ALL)
T (ALL)
T (ALL)
T (ATL)
Pre-B (CML-BC)
Pre-B (ALL)
Pre-B (ALL)
T (ALL)
Pre-B (ALL)
T (ALL)
T (ALL)
Pre-B (ALL)
Pre-B (ALL)
T (CLL)
Pre-B (ALL)
Pre-B (AUL)
Pre-B (ALL)
Myeloma
Myeloma
PCL
PCL
Northernt
PCR+
Cell Line
Myeloma cell lines
MM-l
MM-S1
NCLH929
OPM-2
U-266
U-l996
B-/T-lymphoma cell lines
BJAB
CA-46
DAUDI
DEL
DG-75
DOHH-2
EB-l
HDLM-l
HDLM-2
HDLM-3
HT-58
KARPAS-299
KARPAS-422
KM-H2
L-428
MC-l16
MH-l
NAMALWA-IPN-45
NAMALWA
PFI-285
RL
ROS-l7
ST-4
SUP-T1
U-698"
WIEN-l33
WSU-NHL
WSU-WM
Myeloid leukemia cell lines
EM-2
EM-3
EOL-l
EOL-3
GDM-l
GM-l53
HL-60
HMC-1
KASUMI-1
KCL-22
KG-l
KG-1A
KBM-7
KOPM-28
KU-812
KU-812F
KY-821
KYO-1
MR-87
MOLM-6
MOLM-7
MOLM-8
NB-4
OCI-AML-5
Type/Origin*
Northernt
PCRS
+++
+
+
Myeloma
Myeloma
Myeloma
Myeloma
Myeloma
Myeloma
Burkitt
Burkitt
Burkitt
MH
Burkitt
B-NHL
Burkitt
Hodgkin
Hodgkin
Hodgkin
B-NHL
T-NHL
B-NHL
Hodgkin
Hodgkin
B-NHL
MH
Burkitt
Burkitt
T-NHL
B-NHL
Burkitt
T-NHL
T-NHL
B-NHL
Burkitt
B-NHL
WaldenstrBm
CML-BC
CML-BC
AML-eosino
AML-eosino
CML-BC
AML
AML M2
MCL
AML M2
CML-BC
AML
AML
CML-BC
CML-BC
CML-BC
CML-BC
AML
CML-BC
AML
CML-BC
CML-BC
CML-BC
AML M3
AML
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
CELL c-KIT IN MEGAKARYOBLASTIC
Table 1. Expression of o k i t mRNA in Leukemia-Lymphoma C
e
l Lines (Cont'd)
Type/Origin*
Cell Line
Myeloid leukemia cell lines
PL-21
TI-l
TS9;22
UCSD/AML-1
YS9;22
Monocytic leukemia cell
lines
AML-193
cm-1
DD
JOSK-I
JOSK-K
JOSK-M
JOSK-S
KBM-3
KBM-5
ML-2
MONO-MAC-6
MV4-11
NOMO-1
OCI-AML-1
OCI-AML-2
PLB-985
RC-2A
RW-LEU-4
SCC-3
SKM-1
'
Northernt
PCRS
Cell Line
Monocytic leukemia cell
lines
THP- 1
U-937
AML M3
AML M2
CML-BC
AML
CML-BC
TK-1B
X-376
Erythroid leukemia cell lines
F36P
F36EGM
HEL
K-562
KMOE-02
OCI-M1
OCLM2
TF-l
Megakaryoblastic leukemia
cell lines
CHRF-288-11
CMK
DAM1
LAMA-84
M-07e
MEG-01
MEGAL
MKPL-l
MOLM-1
UT-?
AML M5
AML M5
Histiocytic
lymph.
AML M4
AML M5
CML-BC
AML M5
AML M4
CML-BC
AML M4
AML M5
AML M5
AML M5
AML M4
AML M4
AML M4
AML M4
CML-BC
NHL
AML M5
Northernt
Type/Origin*
PCRS
AML M5
Histiocytic
lymph.
AML M4
AML
-
AML M6
AML M6
AML M6
CML-BC
AML M6
AML M6
AML M6
AML M6
-
AML M7
AML M7
AML M7
CML-BC
AML M7
CML-BC
AML M7
AML M7
CML-BC
AML M7
-
+
(+)
+
+++
++
++
+++
+++
++
++
+++
++
++
+++
++
++
Abbreviations: ATL, adult T-cell leukemia; AUL, acute undifferentiated leukemia; B, B-cell; Burkitt, Burkitt's lymphoma; CLL, chronic lymphocytic
leukemia; CML-BC, chronic myeloid leukemia in blast crisis; HCL, hairy cell leukemia; Hodgkin, Hodgkin's lymphoma; MCL, mast cell leukemia;
MH, malignant histiocytosis; NHL, non-Hodgkin's lymphoma; PCL, plasma cell leukemia; PLL, prolymphocytic leukemia; Pre-B, pre-B-cell; T, Tcell.
Cell lines were assigned t o the respective categories based on their origin and their phenotypic and functional characteristics (eg, surface
markers, expression of hemoglobin, monocyte-specific esterase, etc.); subtypes are given as indicated in the original publications."28
t Intensity of bands on Northern blots: -, negative; (+l, weakly positive; +, ++, +++, various degrees of positivity. Equal loading of the
lanes and integrity of theRNA were verified by ethidium bromide staining of the gel
and rehybridization with &actin.
Results of RT-PCR: -, negative (no band); +, positive.
*
first dissolved in 100% ethanol and then further diluted to the final
concentrations in culture medium. The following growthfactors (all
obtained from Boehringer. Mannheim, Germany) were used at the
same concentrations in all four cell lines: 30 U/mL IL-3, 30 UlmL
GM-CSF, 30 ng/mL IGF-I, and 30 ng/mL IGF-11. M-07e cells were
Fig 1. Northern blot analysis
of c-kit expression in representative examples of positiveand
negative leukemia cell lines is
shown. The specific band has a
size of 5.0 kb. Equal loading of
each lane and integrity of the
RNA ware verifiedin all samples
byethidiumbromidestaining
immediatelyaftergel
electrophoresis and by rehybridization
with a p-actin probe.
commonly cultured in R P M I 1640 medium containing conditioned
medium (10% v01 supernatant) o f the human bladder carcinoma cell
line 5637 that produces IL-3 and GM-CSF. Before the experiments,
the M-07e cells were washed twice with RPMI1640 and starved for
the subsequent 12 hours by withdrawal o f the 5637 cell supernatant.
"
"
"
"
"
"
"
"
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
HU ET AL
2136
Evaluation of Cell Proliferation
Molecular Probes
The methylthiotetrazole (MTT) assay was usedto evaluate the
proliferation of the megakaryoblastic cells. The use of MTT as an
indicator of cell growth has been
well
Cells were
grown in microtiter plates at a density of 2 X IO‘lwell in the presence
ofthe different inducers or growth factors mentioned above. The
cells were pulsed for 4 hours with I O pL MTT solution [5 mglmL 3(4,5-dimethylthiazol-2-yl)-2.5-diphenyltetrazolium
bromide (Sigma)
in phosphate-buffered saline (PBS)]. The reaction was stopped with
120 pL of5% formic acid in isopropanol. The absorbances were
measured at 570 nm with an enzyme-linked immunosorbent assay
reader. Actual cell number counting was also used to measure the
proliferation ofthe cells. Aliquots of cells were taken from each
treatment group at the four time points (24, 48. 72, and 96 hours),
and viable cells were enumerated after exposure to trypan blue.
Results were expressed as percentage of untreated control cells.
The DNA fragments obtained from the recombinant plasmids carrying the gene sequences studied are c-kir (a 1.25-kb Sst I cDNA
fragment cloned in pUCl19; obtained from American Type Culture
Collection [ATCC; Rockville, MD])’ and hamster 0-actin (a 1.25kb Psr I fragment cloned in pBR322).
Morphologic Evaluation
Aggregation and adhesion of the leukemia cells were examined
in the culture wells or flasks under an inverted microscope. Morphologic features were reviewed on cytospin slide preparations stained
with May-Griinwald-Giemsa stain.
RNA Preparation and Northern Blot Analysis
For Northern blot analysis, total RNAwaspreparedusingthe
guanidinium isothiocyanatelCsC1method. Equal quantities of total
RNA (10 pg) were separated on 1.0% formaldehyde agarose gels.
The fractionated RNA was transferred tonylonfilters that were
subsequently cross-linked with UV light. The gels were stained with
ethidium bromide to ensure that equal amounts of RNA were analyzed and that no degradation had occurred. After prehybridization
at 60°C for 2 hours, the RNA was hybridized to nick-translated ”PaCTP-labeled cDNA probes for 18 hours at 60°C. After stringent
washing, the blots were exposed to x-ray films with an intensifying
screen at -80°C. Sizes of mRNAs were estimated from the positions
of 28s and 18s bands in ethidium bromide-stained blot gels. The
same filters were subsequently hybridized with a 0-actin probe.
Immunophenotyping Analysis
RT-PCR
The cells were immunophenotyped using monoclonal antibodies
(MoAbs) directed against the antigens CD13 (MY7; Coulter, Krefeld, Germany), CD14 (FMCI7; Flinders Medical Center, Melbourne,
Australia), CD33 (MY9; Coulter), HLA-DR (RFDR-2; Royal Free
Hospital, London, UK), CD4la (glycoprotein [GP] IlblIIIa; Dako,
Hamburg, Germany), and CD42b (GP Ib; Dako). Briefly, in a microtiter plate system, the cells were washed twice with PBS and then
incubated for 30 minutes with specific MoAbs at optimal dilutions.
All reactions were examined with the indirect method: after incubation with MoAbs and subsequent washing steps, cells were stained
for 45 minutes with fluorescein isothiocyanate-conjugatedgoat antimouse isotype-specific F(ab‘)’ reagents (SBA, Dunn, Asbach,
Germany). The labeled cells were analyzed byflow cytometry
(FACScan; Becton Dickinson, Heidelberg, Germany). In every determination, a negative control of an irrelevant mouse MoAb of the
same isotype as the MoAb under testing was included.
First-strand cDNA was synthesized using
a reverse transcriptase pmamplification system kit (Super Script; GIBCO) following the manufacturer’s instructions. Five micrograms of total RNA as a template were
incubated with 50 ng of random hexamer nucleotidesin a final volume
I O minutes,
of 16 pL DEPC-H20.After heating the mixture at 70°C for
I pL (200 U) of Moloney murine leukemia virus reverse transcriptase,
2 pL IOX synthesis buffer, and 1 pL of IO mmol/L dNTP mix were
added to the reaction system. The reaction mixture was then incubated
at 42°C for 50 minutes, at 90°C for 5 minutes and was then quickly
chilled on ice. After brief centrifugation, 2 U RNase H were added to
the reaction mixture, and this mixture was incubated for 20 minutes at
37°C to digest the template RNA.
Five microliters of the reverse transcriptase reaction mixture containing the first-strand cDNA were diluted with PCR buffer (IOX:
500 mmol/L KCI, 15 mmoVL MgCI2, 100 mmol/L Tris-HCI pH 8.3,
0.01% gelatin) containing 50 pmol ofeach upstream and downstream
B
Actin
.
)
Fig 2. RT-PCR products obtained from a panel of
leukemia-lymphoma celllines are shown. (AI RNA
was reverse transcribed into cDNA that was amplified by PCR using c-kit-specific primers. The 1.1-kb
PCR products were visualized after electrophoresis
by ethidium bromide staining. Whereas 3 lymphoid
leukemia and lymphoma celllines (PC-53, JVM-2,
HDLM-3) were negative, the remaining 9 cell linesof
myeloid, monocytic, erythroid, or megakaryoblastic
origin were clearly positive (see also Table 11. (B1
Amplification of actin as a control for the presence
of amplifiable RNA is shown. The PCR product was
detected by
autoradiography
after
hybridization
with a specific probe.
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
2137
c - m IN MEGAKARYOBLASTIC CELL LINES
primer, 50 nmol of dNTP mix, and 1.25 U of Taq DNA polymerase
(Amersham-Buchler, Braunschweig, Germany). The primers used in
the experiment were designed according to sequence data published
(sequence
previously:' sense 5'-GAGGAGATAAATGGAAAC-3'
nucleotides 1702-1719) and antisense 5"GGTGCTCTCTGAAATCTGC-3' (sequence nucleotides 2817-2799). These two primers encompass the c-kif cDNA sequence encoding the cytoplasmic domain
of the SCF receptor. Oligonucleotide primers were prepared on an
automated DNA synthesizer (Cyclone Plus; Millipore, Eschbom,
Germany). The PCR was then performed with a DNA thermal cycler
(Perkin Elmer Cetus, Heidelberg, Germany) for 30 cycles under the
following conditions: 30 seconds at 90°C for denaturation, 30 seconds at 56°C far annealing, and 2 minutes at 72°C for extension.
The amplified PCR products were electrophoresed in 1.0% agarose
gels, were stained with ethidium bromide, and were observed under
UV light. Negative controls were included in each RT-PCR assay
to exclude false-positive results. RT-PCR on RNA from the same
sample was performed in tandem with the c-kirPCR using the
housekeeping gene, &actin, to show that RNA was intact and amplifiable for the exclusion of false-negative results.
Detection of Apoptotic DNA Degradation
For genomic DNA preparation, 5 X IO6 cells were harvested,
washed twice with PBS (pH 7.3, and incubated with lysis buffer
overnight at 50°C. The samples were extracted twice with an equal
volume of phenol-chloroform-isoamyl alcohol. The DNA was precipitated with 7.5 mom ammonium acetate and 100% ethanol, was
treated with 1 ,ug/mL DNase-free RNase (Sigma) for 1 hour at 3 7 T ,
and was dissolved in buffer at a concentration of 1 ,ugh&. Apoptotic
DNA degradation was examined by electrophoresing the DNA in a
1.6% agarose gel.DNA degradation was visible in the ethidium
bromide-stained gel under UV light as a ladder of about 200-bp
fragments.
RESULTS
Constitutive Expression of c-kit
c-kit mRNA expression detected by Northern blot analysis.
To determine the pattern of c-kit expression in different cell
lineages, we analyzed 164 continuous human leukemia and
lymphoma cell lines using Northern blot analysis (Table 1).
Northern blot analysis of total cytoplasmic RNAshowed
specific signals of 5 kb in size on probing with the human
c-kit cDNA in positive cell lines. c-kit mRNA was notfound
in any cell line of the 54 lymphoid leukemia, 11 myeloma,
or 28 lymphoma cell lines. c-kit mRNA was detected in 10
of 10 megakaryoblastic and in 6 of 8 erythroid leukemia cell
lines; 7 of 29 myeloid and 1 of 24 monocytic leukemia cell
lines were also positive for c-kit. These data indicate that ckit gene expression, as detectable by Northern blotting, is
restricted to leukemia cell lines derived from myeloid, monocytic, erythroid, and megakaryoblastic lineages. Most of the
10 megakaryoblastic leukemia cell lines displayed large
amounts of c-kit mRNA. Fig I shows the results of the
Northern blot analysis of some of the cell lines investigated.
c-kit mRNA expression detected byRT-PCR.
Because
some cell lines might express only minimal amounts of ckir mRNA and Northern blotting might not be sensitive
enough for its detection, we further performed RT-PCR on
a panel of 33 randomly selected cell lines: 1 myeloma and
5 lymphoma cell lines and 5 lymphoid, 6 monocytic, 8 myeloid, 5 erythroid, and 3 megakaryoblastic leukemia cell
lines (Table 1 and Fig 2). c-kit mRNA was not detectable
in any of the lymphoma and lymphoid leukemia cell lines
that were investigated with RT-PCR. However, 4 of 8 myeloid, 3 of 6 monocytic, 5 of 5 erythroid, and 3 of 3 megakaryoblastic cell lines expressed c-kit mRNA in the RT-PCR
analysis. All of the Northern-positive cell lines were also
positive in the c-kit RT-PCR, whereas 8 Northern-negative
cell lines were positive after RT-PCR.
Expression of c-kit on Treatment of Megakaryoblastic
Leukemia Cell Lines CMK, DAMI, M-07e, MKPL-1
Efsects of TPA, Bryo I and ATRA on cell proliferation.
The MTT assay and cell counting were used to investigate
the proliferation-inhibiting effects of P A , Bryo 1, and
ATRA on the four megakaryoblastic leukemia cell lines.
Growth inhibition was already clearly observed after 48
hours and became more pronounced at 72 and 96 hours (Fig
3). Whereas TPA was the most effective reagent, the degree
of inhibition varied between cell lines. Results from the MTT
assay correlated well with the actual cell numbers enumerated by cell counting in a hematocytometer with trypan blue.
Effects of growth factors on cell proliferation. The effects of the cytokines IL-3, GM-CSF, IGF-I, and IGF-I1 on
the growth of the four cell lines were also measured with
the MTT assay and by cell counting. As shown in Fig 3,
both IL-3 and GM-CSF strongly enhanced the proliferation
of M-07e cells. IGF-I and IGF-I1also stimulated the growth
of M-07e cells, albeit to a lesser extent. Only IL-3 and GMCSF increased the proliferation of CMK and MKPL-1,
whereas IGF-I and IGF-I1 were not effective. DAMI cells
did not respond to any of the four growth factors.
Morphologic analysis of induced cell lines.All
four
megakaryoblastic cell lines grow as suspension cultures with
less than 5% of the population adhering to the plastic surface
and extending pseudopods. During the 4-day treatment period, untreated cells did not show any signs of spontaneous
morphologic changes. However, cells treated with the various inducers showed significant morphologic changes in all
four cell lines. Within 4 hours of adding the PKC activators
TPA and Bryo 1 or ATRA to the cell culture, the cell morphology began to change, leading to some significant alterations over the next 2 to 3 days (Fig 4). In TPA- or Bryo
l-treated cultures, more than 50% of the cells adhered to the
surface of the flask and aggregated to form clusters. The
cytoplasm increased, and the cell form became clearly more
irregular. An increased number of giant cells was readily
observed. In about 20% to 50% of CMK, DAMI, and MKPL1 cells, we found polykaryons, some withmorethanfive
nuclei; the increased cytoplasm appeared as ruffled extensions. ATRA-treated cells, however, showed different
changes in morphology. Only few cells adhered to the plastic
surface; the increase of the cytoplasm was less abundant,
but there was a prominent development of cytoplasmic budlike processes; and the blast-like nuclei became more dense.
No morphologic changes were observed in the cultures exposed to L-3, GM-CSF, IGF-I, or IGF-11. The appearance
of large cells with nuclei that are either segmented or fragmented was not caused by poor cell culture conditions because the viabilities were 80% or higher throughout the treatment period.
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
HU ET AL
2138
% of untreated Control
180
A
140
120
104
102
100
BO
BO
40
20
0
T PA
Bryo 1
ATRA
GM-CSF
IL-3
IGF-I
IGF-II
YO of untreated Control
1711
,
l
B9
T PA
Bryo 1
ATRA
GM-CSF
IL-3
IGF-II
IGF-I
Fig 3. Inhibitory effects of the three inducers, TPA, Bryo 1 and ATRA, and stimulatory effects of the four factors, GM-CSF, IL-3, IGF-I and
IGF-II, on the growth of the four megakaryoblastic leukemia celllines, (A) CMK, (B) DAMI, (C) M-07e. and (D) MKPL-1, are shown. Cells (2 x
104/wells) were incubated in t h e presence of the inducers TPA, Bryo 1, or ATRA (at 10 mol/L each) or the growth factors GM-CSF (30 U/
mL), IL-3 130 UlmL), IGF-I 130 ng/mL), and IGF-I1 (30 ng/mL) for 96 hours. Shown are the resultsas percentages of control for each cell line
and treatment at (M) 24, (0)
47, (a)72, and (D) 96 hours. M-07e is an IL-3- or GM-CSF-dependent cell line that was normally cultured with
10% (vol) conditioned medium of the 5637 cell line. Before the experiment, M-07e cells were starved; control M-07e cells were incubated
without IL-3, GM-CSF, or 5637-supernatant. Viabilities in all treatment groups were greater than90% at 48 hours and greater than80% at 96
hours as assessed by trypan blue dye exclusionin all treatment groups.
Apoptotic‘ D N A drgmdation. To test whether apoptosis
was induced by the treatment with the biomodulators, the DNA
from the two cell lines DAM1 and M-07e that had been incubated with TPA, Bryo I , and ATRA (each at lo” m o K ) for
96 hours was separated in an agarose gel. Examination of the
ethidium bromide-stained gel under UV light showed that the
DNA of these samples was intact because a DNA ladder with
200-bp fragments was not visible. In contrast, the control sam-
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
C-KIT
IN MEGAKARYOBLASTIC CELL LINES
2139
% of untreated Control
280
286
280 242
m
240 220
-
200 180 180 -
148
140 120
132
n
-
100
80
eo
40
20
0
Bryo 1
T PA
ATRA
GM-CSF
IGF-I
IL-3
IGF-I1
YO of untreated Control
140
120
100
1
I
148
1
160
n
S2
S1
T PA
Bryo 1
ATRA
GM-CSF
IL-3
IGF-I
IGF-It
Fig 3. (Cont'd).
plc of M-07e cells that were starved for 96 hours by withdrawing the S637 supernatant (containing GM-CSF andIL-3) clearly
displayed the DNA degradation in the characteristic oligonucleosomal pattern (data not shown).
1mmurzo)~)henoty~)inganalysis
.J' induced cell lines. The
surface markerprotilcs of the four megakaryoblastic cell
lines, before and after exposure to TPA, Bryo I , or ATRA,
are summarized in Table 2. Except for the negative M-07c
cells, the cell lines showed the typical mcgakaryoblastic surface markers, GPIlb/IlIa (CD4 1 a) and GP Ib (CD42b). Treat-
ment with TPA or Bryo 1 induced an upregulation of several
in the percentage of
surface antigens.Astrongincrease
CD4la- and CD42b-positivecells wasnoticed in CMK,
CD13,
DAMI, and MKPL-l (Fig S). To acertainextent,
CD14, CD33, and HLA-DR expression was also increased
in CMK,DAMI, and MKPL-Icells aftertreatment with
TPA or Bryo 1 butnot ATRA. ATRA, at I O ' mol/L, did
not have any remarkable effects on the expression of those
surface markers studied. In contrast to the other three megakaryoblasticcelllines, no major changes were seen in the
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
HU ET AL
2140
-
. .
"
.
~~
"
"".
-
p
C
=
m
w
"
."
' .".
D
"7
1,
.F
Fig 4. Morphology of megakaryoblastic cell lines exposed to dflerentinducers on May-Grllnwald-Giemsa-stained cytospin slide preparations (original magnification x 250) are shown. (A) untreated DAMI; (B1 DAM1 with ATRA; (C) DAM1 with Bryo 1; (D) DAM1 with TPA; (€1
untreated MKPL-l; and (F) MKPL-l with ATRA. Cells were exposed t o the inducers (each at lo" mollL) for 96 hours. Note the condensation
of the nuclei, budding and platelet-like segmentationof the cytoplasm in (B, C, F), and the prominentcytoplasmic enlargement in (D).
M-07e immunoprofile throughout the 4-day treatment period
when compared with that for the untreated cells.
Modulation of c-kit gene expression. mRNAwas isolated from the cell lines after treatment with the differentiation inducers TPA, Bryo l , and ATRA or with the growth
factors IL-3, GM-CSF, IGF-I and IGF-11. TPA and Bryo 1
upregulated c-kit expression in all four cell lines, with TPA
being the more effective reagent of the two inducers and
DAMI and MKPL-l being the most responsive cell lines (Fig
6). Only a slight, but reproducible decrease in c-kit mRNA was
noted in the four cell lines treated with ATRA; however, c-kit
wasnot completelydownregulated(Fig 6). IL-3. GM-CSF,
IGF-I, and IGF-I1 did not have any significant effects on the
c-kit expression in the four cell lines (Fig 7).
DISCUSSION
Recently, evidence has been presented that c-kit is
also
involved in the proliferation of human leukemia cells.'"''
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
C-KIT
IN MEGAKARYOBIASTIC CELL LINES
2141
Table 2. Surface Marker Analysis of Four MegakaryoblasticCell Lines Treated With IPA, Blyo 1, or ATRA
CMK
DAM1
MoAb
CD
Con
TPA
Bryo 1
ATRA
Con
TPA
Bryo 1
MW
FMC17
MY9
GP Ilb/llla
GP Ib
RFDR-2
CD13
CD14
CD33
CD4la
CD42b
HLA-DR
81
46
83
31
6
11
94
79
85
71
42
50
86
63
90
71
46
27
87
49
90
10
8
20
84
30
100
61
70
80
100
91
100
94
87
96
90
63
95
87
86
88
M-07e
TPA
ATRA
Con
90
58
100
73
73
90
MKPL-1
Bryo
ATRA
1
42
0
40
0
8
0
57
7
47
0
22
15
41
0
48
0
20
4
27
0
38
0
9
0
Con
TPA
Bryo
ATRA
1
81
28
86
42
0
42
78
52
82
80
29
57
72
21
76
63
28
37
79
17
75
44
9
30
mol/L TPA,Bryo1,orATRAfor
96 hoursand were analyzed by flow cytometry (the figures arepercentageof
Cells were treated with
positive cells). Triplicate experiments produced similar results with deviations of less than 5%. Treatment of other cell lines (eg, myeloid
leukemia cell lines HL-60 and NB-4) did not induce upregulation of CD4la or CD42b.
Abbreviations: Con, control (untreated cells).
There are ample data that the c-kit gene is exclusively expressed in human myeloid andnot lymphoid leukemia
ce~~s.17.19,29-31 c-kit gene expression was reportedly detected
in acute lymphoblastic leukemia (ALL) cells, albeit only in
the particular subgroup of myeloid antigen-positive, immature A L L 3 *
In the present study we found c-kif mRNA expression
only in cell lines with myeloid, monocytic, erythroid, or
megakaryoblastic features. All leukemia or lymphoma cell
lines assigned to lymphoid lineages were negative for c-kit
mRNA, even after using RT-PCR in some of the cell lines
to detect any minimal amounts of mRNA. It is of particular
interest that c-kit message was seen by Northern blotting
and/or RT-PCR in all erythroid-megakaryoblastic cell lines.
The results from those cell lines investigated suggest that
about 50% of the myeloid-monocytic lines express c-kit
mRNA, however in amounts clearly lower than those found
in erythroid-megakaryoblastic cell lines.
A striking finding was the difference in c-kit expression
:ig 5. Expressionof the megakaryoblasticdflerentiation antigens
CD4la (GP Ilb/llla) and CD42b (GP
Ib) on MKPL-1 and
CMKcells b i o r e
(a) and after (b) treatment with 10" mollL TPA or Bryo lfor 72 hours,
as determined by flow cytometric analysis, is shown. (1) CD4la on
MKPL-1 treated with TPA; (2) CD42b. MKPL-1 with TPA; (3) CD4la,
CMK with Blyo 1; and (4) CD42b. CMK with Bryo l.See Table 1 for
percentage of positive cells.
between freshly explanted AML samples, most of which
express c-kit mRNA and pr~tein,'~.'~.~'
and established cell
lines withthe phenotypic characteristics of myeloid cells
confirming a previous, albeit significantly smaller, study on
c-kit expression in cell lines3' Short-term cultivation of primaryAML blasts didnot reduce c-kit expres~ion.~'One
explanation might be that cell lines no longer need to express
c-kit, having become entirely autonomous during establishment of the culture because of the outgrowth of a specific,
independent clone.
After our initial studies that defined the category of cells
expressing constitutively c-kit mRNA and that
established
the highest expression of c-kit in megakaryoblastic cell lines,
we next examined whether c-kit mRNA is upregulated or
downregulated during treatment with known differentiation
inducers or with cytokines. We observed the following effects
inallfourcelllines,CMK,DAMI,M-07e,andMKPL-1,
treatedwith TPA or Bryo 1: arrest of proliferation,distinct
morphologic alterations, and changes in surface marker expression. The most remarkable changes were the increased number
of giant cells with multilobulated or multiple nuclei, the segmentation and budding of the cytoplasm, and the increase in
CD4la- and CD42b-positive cells. The morphologic alterations
of megakaryoblastic cell lines triggered by TPA
or Bryo 1 were
paralleled by increases in c-kit mRNA levels.
ATRA slightly downregulated c-kit expression. The morphologic changes that were seen in ATRA-treated cells, including prominent development of cytoplasmic bud-like processes, do not
necessarily
constitute differentiation,
especially in the face of no change in objective markers of
megakaryocytic differentiation. Thus, from the data obtained, we cannot conclude unequivocally that differentiation
occurred after ATRA treatment; alternatively, effects on the
cell lines by ATRAmay include programmed cell death
(apoptosis) without differentiation or dedifferentiation. However, we excluded apoptotic DNA degradation in the two
cell lines DAMI and M-07e. Therefore, depending on the
type of modulators used and, consequently, the different
signal transduction pathways, c-kit expression is augmented
or reduced. The close relationship between erythroid and
megakaryoblastic cell lines is underlined by the shared c-kit
expression and by the fact that many of these cell lines
carry markers of both lineages (eg, hemoglobin and C D 4 l d
CD42b) or can be induced to mature along either pathway
depending on the stimulus applied.
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
HU ET AL
2142
“
”
”
”
”
*
I*
@
I
,
actin
+
Treatment with the cytokines, IL-3, GM-CSF, IGF-I, and
IGF-11, increased cell proliferation significantly in M-07e
cells but only marginally in the other three megakaryoblastic
cell lines that normally grow independent of any growth
factors. During exposure to these cytokines, c-kitmRNA
levels did not change significantly. Further studies on primary cell material that could be more responsive to exogenous cytokines might be veryinformative for elucidating any
potential role of c-kit in the regulation of megakaryoblastic
proliferation. Tumor necrosis factor-cy enhanced c-kit mRNA
expression inall specimens of primary AML (cases were
classified as M 1, M2 and M4,butno cases of M7 were
studied).29These investigators did not find any positive effects of other cytokines on c-kit mRNA levels in AML cells,
such as IL-6, GM-CSF and M-CSF.
This is the first thorough study on the expression of c-kit
in a large panel of well-characterized leukemia cell lines. A
previous report addressed the expression of SCF mRNA and
the mitogenic response to SCF protein in some 3 1 cell lines,
most of which were also analyzed in the present report.”
9
IL-3
Fig 6. c-kit mRNA expression is shown
the in
meaakarvoblastic leukemia cell line MKPL-1 treated
with ATRA, TPA, or Bryo 1. Cells were incubated with
the reagents (each at 10” mol/L) for the timeperiods
indicated on top of the lanes. The specific band has
a sizeof 5.0 kb. Note the downregulation of c-kit
mRNA by ATRA and the increase in c-kit mRNA induced by TPA and Bryo 1. Subsequent hybridization
with actin confirmed the equal loading of the lanes.
I
.
Using RT-PCR, SCF mRNA was detected in 17 of 26 cell
lines (7 of 9 myeloid, 6 of 11 monocytic, 3 of 4 erythroid,
and 1 of 2 megakaryoblastic cell lines according to our nomenclature). These data, together with the demonstration of
SCF receptors on some cell lines, indicated the possibility
of autocrine mechanisms in the growth of these cell lines.
Only 5 of 27 cell lines showed a significant proliferative
response to exogenously added SCF (2 of 9 myeloid, 0 of
11 monocytic, 2 of 4 erythroid, and 1 of 3 megakaryoblastic
cell lines). It was concluded that the cells might already
be maximally activated by the SCF present in FCS or by
endogenously produced SCF. Interestingly, 4 of 5 SCF-responsive cell lines are growth factor-dependent cultures.
Indeed, in these experiments, SCF synergized withIL-3,
GM-CSF, and erythropoietin in inducing proliferation in the
responsive cell lines.’y
In conclusion, our extended survey of leukemia-lymphoma cell lines representing allcell lineages showed the
complete lack of c-kit mRNA expression in lymphoid leukemia or lymphoma cells. However, 15% and 50% of cell lines
IGF-I
IGF- I I
GM-CSF
”
“
”
”
”
”
”
”
Fig 7. Expression
of
c-kit
mRNA is shown in themegakarvobladic leukemia cell line. M07e. exposed tothe
growth
factors,IL-3,IGF-l,IGF-ll,
and
GM-CSF, for the timeperiods indicated on top of each lane.
actin
+
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
C-KIT
IN MEGAKARYOBLASTICCELLLINES
with myeloid-monocytic characteristics were c-kit -Apositive in Northern blot and RT-PCR analysis, respectively.
All erythroid and megakaryoblastic cell lines constitutively
expressed c-kit. Treatment of megakaryoblasts with P A or
Bryo 1 was paralleled by increased c-kit mRNA levels; ckit expression was downregulated in ATRA-treated cells.
The present report used secondary, immortalized leukemia
cell lines as material; certainly, not all data obtained from
cell lines can be extrapolated to primary leukemia cells or
even to normal cells of the same lineage with the same
proliferative and maturational status. Nevertheless, our investigations might further contribute to the understanding
of the role of c-kit in leukemia cells, in general, and in
megakaryoblastic cells, in particular. Further investigations
on the expression and modulation of the SCFlc-kit interaction are expected to elucidate the potential clinical use of
this factor or its inhibition.
ACKNOWLEDGMENT
We thank the scientists who kindly provided the cell lines used
in this study.
REFERENCES
1. Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu
S , Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A: Human
proto-oncogene c-kit: A new cell surface receptor tyrosine kinase
for an unidentified ligand. EMBO J 63341, 1987
2. Nocka K, Tan JC, Chiu E, Chu TY, Ray P, Traktman P, Besmer
P: Molecular bases of dominant negative and loss of mutations at
the murine c-kivwhite spotting locus: W37, Wv, W41 and W.EMBO
J 9:1805, 1990
3. Chabot B, Stephenson DA, Chapman VM, Besmer P,Bemstein
A: The proto-oncogene c-kit encoding a transmembrane tyrosine
kinase receptor maps to the mouse W locus. Nature 335:88, 1988
4. Ogawa M, Matsuzaki Y, Nishikawa S , Hayashi SI, Kunisada
T, Sudo T, Kina T, Nakauchi H, Nishikawa SI: Expression and
function of c-kit in hemopoietic progenitor cells. J Exp Med 174:63,
1991
5. Nocka K, Majumber S , Chabot B, Ray P,Cervone M, Bemstein
A, Besmer P Expression of c-kit gene products in known cellular
targets of W mutations in normal and W mutant mice. Evidence for
an impaired c-kit kinase in mutant mice. Genes Dev 3:816, 1989
6. Zsebo KM, Williams DA, Geissler EN, BroudyVC, Martin
FH, Atkins HL, Hsu RY, Birkett NC, Okino KH, Murdock DC,
Jacobsen F W , Langley KE, Smith KA, Takeishi T, Cattanach BM,
Galli SJ, Suggs SV: Stem cell factor is encoded at the SI locus of
the mouse and is the ligand for the c-kit tyrosine kinase receptor.
Cell 63:213, 1990
7. Huang E, Nocka K, Beier DR, Chu TY, Buck J, Lahm HW,
Wellner D, Leder P, Besmer P: The hematopoietic growth factor
KL is encoded by the SI locus and is a ligand of c-kit receptor, the
gene product of the W locus. Cell 63:225, 1990
8. Williams DE, Eisenman J, Baird A, Ranch C, Van Ness K,
Manch CJ, Park LS, Martin U, Mochizuki DY, Boswell HS, Burgess
GS, Cosman D, Lyman S D Identification of a ligand for the c-kit
proto-oncogene. Cell 63:167, 1990
9. Okada S , Nakauchi H, Nagayoshi K, Nishikawa S , Nishikawa
SI, Miura Y, Suda T: Enrichment and characterization of murine
hematopoietic stem cells that express c-kif molecule. Blood 78: 1706,
1991
10. Carow CE, Hangoc G , Cooper SH, Williams DE, Broxmeyer
HE: Mast cell growth factor (c-kif ligand) supports the growth of
human multipotential progenitor cells with a high replating potential.
Blood 78:2216, 1991
2143
11. Zsebo KM, Wypych J, McNiece IK, Lu HS, Smith K A , Kwhare SB, Sachdev RK, Yuschenkoff VN, Birkett NC, Williams LR,
Satyagal V N , Tung W, Bosselman RA, Mendiaz EA, Langley KE:
Identification, purification and biological characterization of hematopoietic stem cell factor from buffalo rat liver-conditioned medium.
Cell 63:195, 1990
12. managan JG, Leder P The kit ligand: A cell surface molecule
altered in Steel mutant fibroblasts. Cell 63:185, 1990
13. McNiece IK, Langley KE, Zsebo KM: Recombinant human
stem cell factor synergizes with GM-CSF, G-CSF, L - 3 and Epo to
stimulate human progenitor cells in the myeloid and erythroid lineages. Exp Hematol 19:226, 1991
14. de Vries P, Brasel KA, Eisenman JR, Alpert AR, Williams
DE: The effect of recombinant mast cell growth factor on purified
murine hematopoietic stem cells. J Exp Med 173:1205, 1991
15. Metcalf D, Nicola NA: Direct proliferative actions of stem
cell factor on murine bone marrow cells in vitro: Effects of combination with colony-stimulating factors. Proc NatlAcad
Sci USA
88:7420, 1991
16. Tohda S , Yang GS, Ashman LK, McCulloch EA, Minden
MD: Relationship between c-kit expression and proliferation in acute
myeloblastic leukemia cell lines. J Cell Physiol 154:410, 1993
17. Lerner N B , Nocka KH, Cole SR, Qiu FH, Strife A, Ashman
LK, Besmer P: Monoclonal antibody YB5.B8 identifies the human
c-kit protein product. Blood 77:1876, 1991
18. Kuriu A, Ikeda H, Kanakura Y, Griffin JD, Druker B, Yagura
H, Kitayama H, Ishikawa J, Nishiura T, Kanayama Y, Yonezawa
T, Tarui S: Proliferation of human myeloid leukemia cell line associated with the tyrosine-phosphorylation and activation of the protooncogene c-kit product. Blood 78:2834, 1991
19. Pietsch T, Kyas U, Steffens U, Yakisan E, Hadam MR, Ludwig WD, Zsebo K, Welte K: Effects of human stem cell factor (ckit ligand) on proliferation of myeloid leukemia cells: Heterogeneity
in response and synergy with other hematopoietic growth factors.
Blood 80: 1199, 1992
20. Sato T, Fuse A, Eguchi M, Hayashi Y, Ryo R, Adachi M,
Kishimoto Y, Teramura M, Mizoguchi H, Shima Y, Komori I, Sunami S , Okimoto Y, Nakajima H: Establishment of a human leukaemic cell line (CMK) with megakaryocytic characteristics from a
Down’s syndrome patient with acute megakaryoblastic leukemia. Br
J Haematol 72: 184, 1989
21. Greenberg SM, Rosenthal DS, Greeley TA, Tantravahi R,
Handin RI: Characterization of a new megakaryocytic cell line: The
Dami cell. Blood 72:1968, 1988
22. Avanzi GC, Lista P, Giovinazzo B, Miniero R, Saglio G,
Benetton G , Coda R, Cattoretti G, Pegoraro L: Selective growth
response to IL-3 of a human leukaemic cell line with megakaryoblastic features. Br J Haematol 69:359, 1988
23. Takeuchi S, Sugito S, Uemura Y, Miyagi T, Kubonishi I,
Taguchi H, Enzan H, Ohtsuki Y, Miyoshi I: Acute megakaryoblastic
leukemia: Establishment of a new cell line (MKPL-I) in vitro and
in vivo. Leukemia 6:588, 1992
24. Mosmann T: Rapid colorimetric assay for cellular growth
and survival: Application to proliferation and cytotoxicity assays. J
Immunol Methods 6555, 1983
25. Monner DA:An assay for growth of mouse bonemarrow
cells in microtiter liquid culture using the tetrazolium salt MTT, and
its application to studies of myelopoiesis. Immunol Letters 19:261,
1988
26. Uphoff CC, Gignac SM, Metge K, Zschunke F, Radzun HJ,
Drexler HG: Expression of the monocyte-specific esterase gene in
leukemia-lymphoma cell lines. Leukemia 758, 1993
27. Drexler HG, Gaedicke G , Minowada J: Isoenzyme studies in
human leukemia-lymphoma cell lines. I. Carboxylic esterase. Leuk
Res 9:209, 1985
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
21 44
28. Drexler HG, Minowada J: Lymphocytic leukemia and
lymphomas, in Herberman RB, Mercer CW (eds): Immunodiagnosis
of Cancer (ed 2). New York, NY, Marcel Dekker, 1990, p 243
29. Brach MA, Biihring HJ, &R HJ, Ashman LK, Ludwig WD,
Mertelsmann RH, Henmann F: Functional expression of c-kit by
acute myelogenous leukemia blasts is enhanced by tumor necrosis
factor-a through posttranscriptional mRNA stabilization by a labile
protein. Blood 80:1224, 1992
30. Ashman LK, Roberts MM, Gadd SJ, Cooper SJ, Juttner CA:
Expression of a 150 kD cell surface antigen identified by monoclonal
HU ET AL
antibody YB5.B8is associated withpoor prognosis in acute nonlymphoblastic leukemia. Leuk Res 12:923, 1989
31. WangC, Curtis JE, Geissler EN, McCulloch EA, Minden
MD: The expression of the proto-oncogene c-lat in the blast cells
of acute myeloblastic leukemia. Leukemia 3:699, 1989
32. Nishii K, Kita K, Miwa H, Kawakami K, Nakase K, Masuya
M, Morita N, Omay SB, Otsuji N, Fukumoto M, Shirakawa S: ckit gene expression in CD7-positive acute lymphoblastic leukemia:
Close correlation with expression of myeloid-associated antigen
CD 13. Leukemia 6:662, 1992
From www.bloodjournal.org by guest on October 2, 2016. For personal use only.
1994 83: 2133-2144
c-kit expression in human megakaryoblastic leukemia cell lines
ZB Hu, W Ma, CC Uphoff, H Quentmeier and HG Drexler
Updated information and services can be found at:
http://www.bloodjournal.org/content/83/8/2133.full.html
Articles on similar topics can be found in the following Blood collections
Information about reproducing this article in parts or in its entirety may be found online at:
http://www.bloodjournal.org/site/misc/rights.xhtml#repub_requests
Information about ordering reprints may be found online at:
http://www.bloodjournal.org/site/misc/rights.xhtml#reprints
Information about subscriptions and ASH membership may be found online at:
http://www.bloodjournal.org/site/subscriptions/index.xhtml
Blood (print ISSN 0006-4971, online ISSN 1528-0020), is published weekly by the American
Society of Hematology, 2021 L St, NW, Suite 900, Washington DC 20036.
Copyright 2011 by The American Society of Hematology; all rights reserved.