NSP k2 k1 k3 KF KF N I U - Web Hosting at UMass Amherst
advertisement

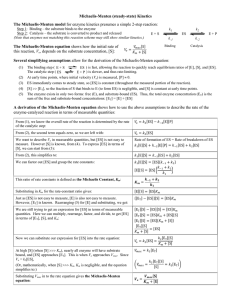
Symposium on Protein Misfolding Diseases Biochemistry & Molecular Biology website May 1-2, 2007 University of Massachusetts Amherst Tuesday, May 1 1:00-1:30pm Registration 1:30-3:30pm Panel discussion: Therapeutic approaches to protein misfolding diseases Moderator: Jonathan King, MIT Panel members: Mark Findeis, Satori Pharmaceuticals; Richard Labaudinaire, FoldRx; Pedro Huertas, Harvard-MIT Division of Health Sciences and Technology; Tim Edmunds, Genzyme 4 pm Special Seminar by Jeffery Kelly, Scripps; "Understanding and ameliorating age onset neurodegenerative diseases“ (A student-invited seminar hosted by the Chemistry-Biology Interface Program) Wednesday, May 2 8:30-9:00am Registration 9-9:50am Valina Dawson, Johns Hopkins; "Proteins behaving badly: Clues to Parkinson's disease" 9:50-10:40am Ron Wetzel, Univ. of Pittsburgh; "The long and short of neurotoxic polyglutamine sequences" 11:00-11:50am Rick Morimoto, Northwestern; "Stress and misfolded proteins: Insights into mechanisms of aging and neurodegenerative disease" 11:50-2:00pm Lunch/posters 2:00-2:50pm Ron Kopito, Stanford; "The ubiquitin system in protein homeostasis" 2:50-3:40pm Byron Caughey, Rocky Mtn. Labs.; "Prion protein aggregation and disease" 4:00-5:00pm Susan Lindquist, the 2007 John Nordin Lecturer, Whitehead Inst.; "Yeast as a discovery platform for protein folding diseases" 5:00-6:00pm Reception The symposium will take place in the Murray D. Lincoln Campus Center at the University of Massachusetts Amherst in the Pioneer Valley of Western Massachusetts. N KFI-N k-2 k2 I k3 KFU-I k-1 k1 U NSP © 2004 New Science Press Ltd new-science-press.com In association with BioMed Central 1 Catalysis and control of protein function Structure and mechanism in protein science. A. Fersht. Chapter 3A and 3B. Petzco & Ringe Chapter 2.6 to 2.9. The concept of steady state In dynamic systems Voet Biochemistry 3e Page 478 © 2004 John Wiley & Sons, Inc. [S0] >> [ET] pre-steady state 2 Michaelis-Menten Mechanism k1 E+S ES k2 P + E k-1 v0 = k2 [ES] = k2 [E]T [S] / (KM + [S]) = Vmax [S] / (KM + [S]) KM is analogous to the dissociation constant of the Michaelis complex k << k (rapid equilibrium) 2 -1 KM = k-1/k1 = KdS At low [S] At high [S] v ∝ [S] v = Vmax = ET kcat Michaelis-Menten Equation The equation holds for many mechanisms, not only for the M-M mechanism. (e.g. Briggs-Haldane kinetics: KM > KdS) v0 = Vmax [S] / (KM + [S]) Vmax = kcat [ET] 3 The significance of the Michaelis-Menten parameters • kcat : the catalytic constant – Is a first order rate constant that refers to the properties and reactions of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes. – Often called the “turnover number” The significance of the Michaelis-Menten parameters • KM : real and apparent equilibrium constant – Only KM = KdS for the simple M-M mechanism – The KM is an apparent dissociation constant that may be treated as the overall dissociation constant of all enzyme-bound species – In all cases is the substrate concentration at which v = Vmax/2 4 The significance of the Michaelis-Menten parameters • kcat/KM : the specificity constant – kcat/KM relates the reaction rate to the concentration of free enzyme, rather than total enzyme. – The kcat/KM is an apparent second-order rate constant that refers to the properties and the reactions of the free enzyme and free substrate – v = [E] [S] kcat/KM Mechanism of regulation of protein function • Why? – To ensure that the protein is only present in its active form in the specific compartment where is needed • How? – Signal sequence – Attachment of lipid tail that insert into membranes – Structural interaction domain (e.g., recognize phosphorylation in other protein) • Localization is a dynamic process (e.g. transcription factors) • When the protein is not in the location where it is needed, very often it is maintained in an active conformation. 5 Protein activity can be regulated by binding of an effector • Binding of effector molecules can induce conformational changes that produce inactive or active forms of the protein. • Effectors may bind noncovalently or may modify the covalent structure of the protein, reversibly or irreversibly. • Often, a product of an enzyme reaction can act as a competitive inhibitor. • Ligands, including reaction products, may also bind to sites remote from the active site (allosteric regulation). S P conc. in cell 10 nM X P Kd of allosteric regulator = 0.1 μM active P inactive Thermodynamic and kinetics of disulfide bond formation in proteins SH P S + GSSG SH + 2 GSH P S K = [P(SS)]eq [GSH]eq2 / [P(SH)2]eq [GSSG]eq Thermodynamics Rate of oxidation = k [P(SH)2] [GSSG] Kinetics You have 2 different solutions containing: a) 0.1 mM GSH and 1 mM GSSG b) 1 mM GSH and 100 mM GSSG How is the concentration of the oxidized and reduced protein at equilibrium in each tube? If you add the reduced protein to each tube, which one will have the highest initial rate of oxidation ? 6