TIME-LAPSE SCREENING BY PARALLELIZED LENSFREE IMAGING
advertisement
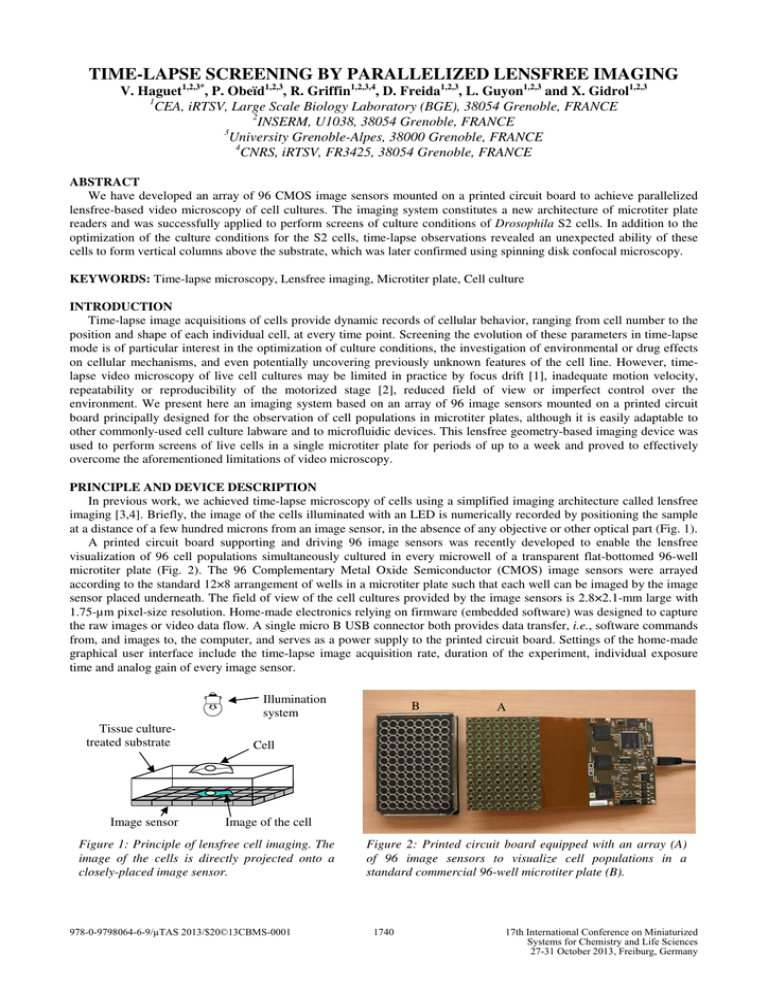
TIME-LAPSE SCREENING BY PARALLELIZED LENSFREE IMAGING V. Haguet1,2,3*, P. Obeïd1,2,3, R. Griffin1,2,3,4, D. Freida1,2,3, L. Guyon1,2,3 and X. Gidrol1,2,3 1 CEA, iRTSV, Large Scale Biology Laboratory (BGE), 38054 Grenoble, FRANCE 2 INSERM, U1038, 38054 Grenoble, FRANCE 3 University Grenoble-Alpes, 38000 Grenoble, FRANCE 4 CNRS, iRTSV, FR3425, 38054 Grenoble, FRANCE ABSTRACT We have developed an array of 96 CMOS image sensors mounted on a printed circuit board to achieve parallelized lensfree-based video microscopy of cell cultures. The imaging system constitutes a new architecture of microtiter plate readers and was successfully applied to perform screens of culture conditions of Drosophila S2 cells. In addition to the optimization of the culture conditions for the S2 cells, time-lapse observations revealed an unexpected ability of these cells to form vertical columns above the substrate, which was later confirmed using spinning disk confocal microscopy. KEYWORDS: Time-lapse microscopy, Lensfree imaging, Microtiter plate, Cell culture INTRODUCTION Time-lapse image acquisitions of cells provide dynamic records of cellular behavior, ranging from cell number to the position and shape of each individual cell, at every time point. Screening the evolution of these parameters in time-lapse mode is of particular interest in the optimization of culture conditions, the investigation of environmental or drug effects on cellular mechanisms, and even potentially uncovering previously unknown features of the cell line. However, timelapse video microscopy of live cell cultures may be limited in practice by focus drift [1], inadequate motion velocity, repeatability or reproducibility of the motorized stage [2], reduced field of view or imperfect control over the environment. We present here an imaging system based on an array of 96 image sensors mounted on a printed circuit board principally designed for the observation of cell populations in microtiter plates, although it is easily adaptable to other commonly-used cell culture labware and to microfluidic devices. This lensfree geometry-based imaging device was used to perform screens of live cells in a single microtiter plate for periods of up to a week and proved to effectively overcome the aforementioned limitations of video microscopy. PRINCIPLE AND DEVICE DESCRIPTION In previous work, we achieved time-lapse microscopy of cells using a simplified imaging architecture called lensfree imaging [3,4]. Briefly, the image of the cells illuminated with an LED is numerically recorded by positioning the sample at a distance of a few hundred microns from an image sensor, in the absence of any objective or other optical part (Fig. 1). A printed circuit board supporting and driving 96 image sensors was recently developed to enable the lensfree visualization of 96 cell populations simultaneously cultured in every microwell of a transparent flat-bottomed 96-well microtiter plate (Fig. 2). The 96 Complementary Metal Oxide Semiconductor (CMOS) image sensors were arrayed according to the standard 12×8 arrangement of wells in a microtiter plate such that each well can be imaged by the image sensor placed underneath. The field of view of the cell cultures provided by the image sensors is 2.8×2.1-mm large with 1.75-µm pixel-size resolution. Home-made electronics relying on firmware (embedded software) was designed to capture the raw images or video data flow. A single micro B USB connector both provides data transfer, i.e., software commands from, and images to, the computer, and serves as a power supply to the printed circuit board. Settings of the home-made graphical user interface include the time-lapse image acquisition rate, duration of the experiment, individual exposure time and analog gain of every image sensor. Illumination system Tissue culturetreated substrate Image sensor B A Cell Image of the cell Figure 1: Principle of lensfree cell imaging. The image of the cells is directly projected onto a closely-placed image sensor. 978-0-9798064-6-9/µTAS 2013/$20©13CBMS-0001 Figure 2: Printed circuit board equipped with an array (A) of 96 image sensors to visualize cell populations in a standard commercial 96-well microtiter plate (B). 1740 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences 27-31 October 2013, Freiburg, Germany RESULTS AND DISCUSSION The 96-imager electronic board was first applied to determining appropriate culture conditions for the S2 cell line derived from the Drosophila melanogaster embryo (Fig. 3). These 7µm-large cells have been reported as displaying semi-adherent behavior with a tendency to form aggregates after several hours of culture. We adapted published culture conditions to standard 96-well microtiter plates (Falcon tissue culture plates from Corning Life Sciences, Hazebrouck, France). Plates were seeded with S2 cells in Schneider's Drosophila medium containing 10% heat-inactivated fetal bovine serum (Invitrogen, Saint-Aubin, France). The 96 image-series showing proliferation and aggregation processes, as well as motility, of the S2 cells were recorded over 125 h, starting from initial densities between 103 and 7.5×104 cells/well (Fig. 3A). The unique design feature of capturing repetitive wide-field views of 2.8×2.1 mm increases the likelihood of detecting even infrequent cell events; moreover, the highly-parallelized imaging capabilities of the 96-imager electronic board let each experimental condition be replicated in several wells of the microtiter plate. Together, these factors enhance statistically significant analysis and interpretation of quantitative data collected over the entire time range. By culturing S2 cells at various densities in similar conditions, we were able to optimize conditions for S2 cell culture in a single step showing, for example, that cultures with an initial density of 1000 cells/well were too dilute to sustain S2 cell proliferation, while those at ≥5.0×104 cells/well were already confluent. In week-long experiments, we observed absolutely no drift in imaging position (Fig. 3C), which makes this imaging device very competitive for time-lapse acquisition in comparison to conventional video microscopy equipment. 1000 A 1 5000 2 3 10,000 4 5 25,000 6 7 8 50,000 9 75,000 cells/well 10 11 12 A B C D E F G H B t0 C t0 t0+24h t0+48h t0+100h t0+125h 250 µm t0+72h 250 µm Figure 3: Time-lapse screening by parallelized lensfree imaging. (A) Global view at t0+48h of the 96 cultures of S2 cells seeded at 6 different cell densities. (B) Whole field of view of S2 cell culture showing 1185 cells (initial cell density 10,000 cells/well). The contrast of the entire image was enhanced by median filtering to rectify inhomogeneous illumination of the culture. (C) Zoom patches (showing 15% of the whole raw images) at 6 time points as indicated during time-lapse microscopy of S2 cells for 125 h (i.e., 5 days 5 h). Note proliferation and 3D aggregation of S2 cells. White arrow: small dust particle present under the microtiter plate whose stable position over the entire time range testifies to the absence of drift in imaging position. As an example of new cellular phenomena which can be uncovered using time-lapse microscopy, we have observed a tendency for the S2 cells to form vertical columns above the substrate, a behavior not previously detected by conventional microscopy methods. In fact, lensfree imaging creates a characteristic optical signature obtained when two or more S2 cells are superimposed: the resulting assembly has a darker contour and frequently more diffraction fringes than single S2 cells (Fig. 4A). The number of supplementary diffraction fringes is indicative of the number of superimposed cells. The S2 cell columns can last several hours or days, laterally move above the substrate surface like single cells, and sometimes disassemble. Besides successive addition of cells, columns can also be created by mitosis occurring in the vertical 1741 direction, the superimposition of the daughter cells being detected by the same optical signature as the columns. In some cases, the columns of daughter cells were observed to disassemble within a few minutes or hours. Classical cell divisions along the substrate plane were also observed. We assume the semi-adherent S2 cells vertically aggregate in order to minimize contact with the surface. This unexpected behavior of column formation by S2 cells was independently confirmed by spinning disk confocal microscopy (Fig. 4B-E). For fluorescence images, the cell nuclei were labeled for 10-15 min with vital dye Hoechst 33258 (Molecular Probes Europe, Leiden, The Netherlands) initially at 2 mg/mL, diluted 1/5000. Samples were washed with medium and deposited onto the microtiter plates. Optical slices of the cells were made every 500 nm using a spinning disk microscope Eclipse Ti from Nikon (Champigny-sur-Marne, France) with a 20× objective and a 1.5× lens (approx. magnification of 30×). Phase contrast observations of superimposed S2 cells (Fig. 4B) are in full agreement with similar assemblies of the cell nuclei as imaged by fluorescence microscopy (Fig. 4C,E). A B Cell 1 (9 µm) Cell 1 Cell 3 5 µm Cell 4 Z Cell 3 Cell 1 1300 Intensity (a.u.) E Cell 2 Cell 3 Cell 4 Cell 4 D Slice 30 Cell 3 (14.5 µm) Cell 2 (top) C Slice 19 Cell 2 Cell 1 Cell 3 Cell 2 1100 Cell 4 900 700 500 0 5 10 15 20 25 Height Z (µm) Figure 4: (A) Lensfree imaging of S2 cells forming a column. At about t0+36h the cell C divides along the substrate plane into daughter cells D and E. At t0+50h the cells B and D form a 2-cell column F (E moves away) which then joins the cell A and creates a 3-cell column G at t0+55h20. (B-E) Spinning disk confocal microscopy of two columns of two S2 cells each. (B) Phase contrast image, (C) fluorescence images and (D) schematic representation of the columns. (E) Stacking of the cells deduced from the fluorescence intensities of their nuclei. Kinetics of cell-substrate interactions were further investigated by screening S2 adhesion and motility on various substrate materials every 30 min for 170 h (i.e., 7 days 2 h). Matrigel, collagen, fibronectin, untreated glass, oxygen plasma-treated glass and polystyrene samples were deposited in a 12-well microtiter plate which was then seeded with S2 cells at 105 cells/mL and positioned on the 96-imager electronic board. The 5 or 6 image sensors present under each well of the 12-well microtiter plate allow characterization of several positions in the cell cultures without any motion of stage or objective. We observed higher proliferation, motility and aggregation rates for S2 cells on Matrigel, fibronectin or polystyrene than on the other materials. CONCLUSION We present a new architecture of microtiter plate reader based on parallelized lensfree imaging. Ninety-six 2.8×2.1mm image-series are acquired by an array of 96 image sensors mounted on a printed circuit board. This imaging system provides a physical support for extended time-lapse screens in which the effects of different treatments on complex and dynamic behaviors can be systematically compared. ACKNOWLEDGEMENTS D. Thévenon at CEA is acknowledged for supplying the S2 cell line. Financial support was provided by the National Research Agency (grant ANR-12-EMMA-0051-01). REFERENCES [1] M.M. Frigault, J. Lacoste, J.L. Swift and C.M. Brown, J. Cell Sci., vol. 122, pp. 753-767, March 2009. [2] T. Bartholomäus and B. Espinzoa, Photonics Spectra, pp. 60-61, March 2009. [3] M. Gabriel, V. Haguet, N. Picollet-D’hahan, M. Block, B. Fouqué and F. Chatelain, Proc. µTAS, Oct. 12-16, 2008, San Diego, USA, vol. 2, pp. 1600-1602. [4] M. Gabriel, N. Picollet-D’hahan, M. Block and V. Haguet, Proc. µTAS, Nov. 1-5, 2009, Jeju, South Korea, vol. 1, pp. 278-280. CONTACT *V. Haguet, tel: +33-4-38-78-23-86; vincent.haguet@cea.fr 1742