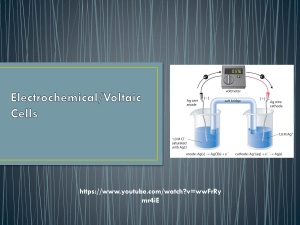
Ph.D. Thesis
advertisement

Development of a Miniaturized Electro-Fluidic Detector for Medical Diagnostics Ph.D. Student: Supervisors: Luca Ianeselli Alessandro Laio Loredana Casalis Sector: Statistical and Biological Physics Academic Year: 2012 / 2013 Contents Abstract . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1 1 Introduction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3 Differential Capacitance . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8 2 Materials and Methods . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11 2.1 Materials . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11 2.2 Device Fabrication . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 13 2.3 Experimental Setup . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 18 2.4 Instruments: Electrical and Topographic Characterization . . . . . . . . . . . . . . . . . . . . . . . . . . . 20 2.5 3 4 2.4.1 DSO Lock-In Amplifier: Model SR830 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20 2.4.2 Heka Bipotentiostat PG 340 usb . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22 2.4.3 Atomic Force Microscope: MFP 3D . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 24 Experimental Procedures: SAM Formation and Hybridization Procedure . . . . . . . . . . . . . . 25 Two-Electrode Configuration . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27 3.1 Electrical Characterization . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27 3.2 DNA-Hybridization . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31 3.3 Time Behavior of the Two-Electrode Setup . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33 3.4 Protein Detection . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 39 3.5 Problems of the Two-Electrode Setup and Possible Solutions . . . . . . . . . . . . . . . . . . . . . . . . 43 Three-Electrode Configuration . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 45 4.1 Electrical Characterization . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 45 4.2 DNA Hybridization as a Function of the Applied Bias Potential . . . . . . . . . . . . . . . . . . . . . . 49 4.3 Time Behavior of the Three-Electrode Setup . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 54 4.4 Calibration Curve of the Three-Electrode Detectors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 62 4.5 Kinetics of DNA Hybridization . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 64 4.6 Passivation with Molecules of Thiolated Ethylene Glycol . . . . . . . . . . . . . . . . . . . . . . . . . . . 67 4.7 Major Achievements of the Ph.D. Work . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 68 VI 5 A Contents Conclusion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 71 5.1 Where we are . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 71 5.2 Perspectives . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 73 Differential Capacitance at the Electrode-Electrolyte Interface: Models of the Differential Capacitance . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 77 B Instruments and Lithographic Procedures . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 81 B.1 Spin-coater . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 81 B.2 Mask Aligners . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83 B.3 e-beam Evaporator . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 85 B.4 Lithografic Procedures . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86 References . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 87 Acknowledgements . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 93 Abstract In the last decades the interest towards personalized therapies has fostered a big number of studies dedicated to the realization and the optimization of bio-detectors to be used as fast diagnostic tools during medical treatment [1, 2, 3, 4]. Among the proposed devices the best performances, both in terms of multiplexing and cost reduction, are expected by the detectors based on electrical readout. These sensors can be integrated with microfluidic networks in the so called Lab − on − a −Chip systems and offer the possibility to develop complete diagnostic kits for the use as a medical practitioner’s bench tool and, ultimately, for rapid and reliable analysis in low-resource areas and in the developing world [5, 6]. In this framework we focused on the development of an electrochemical biosensor based on capacitance readout, for the detection of biomolecules in small sample volumes. We performed electrochemical impedance spectroscopy (EIS) measurements of DNA-hybridization and protein-protein interaction in electrochemical cells with microfabricated gold electrodes. The time stability of the device was tested in two different configurations: two microelectrodes in a microfluidic channel; two microelectrodes plus a reference electrode in an electrochemical cell. Our results demonstrate that the three-electrode setup is more stable, more reproducible, and suitable for real-time measurements. A thorough study of the immobilization strategy of the DNA-molecules on the gold electrodes was carried out. In the last part of the work we performed a test study of DNA-hybridization in real time and we showed that the three-electrode configuration can measure the process in-situ. 1 Introduction Recent trends in biomedicine highlight the importance of accurate and continuous monitoring of disease bio-markers during medical treatment [1, 2, 3, 4]. This emerging field is called personalized medicine and requires the development of new, faster and more reliable detectors for bio-marker recognition, fulfilling the need for point-of-care medical diagnostic tools. Moreover, these devices should have the capability of multiplexing, that is the possibility to combine, in a single analysis, the detection of several bio-markers. Multiplexed analysis of bio-markers leads to more informative results and can thus reduce the numbers of false-positives, increasing the reliability of the analysis [7]. In 2009 Dr. Fouzia Bano, a former Ph.D. student of Sissa who worked in the Nano Innovation Laboratory (NIL) at Elettra, published a paper that goes in this direction [8]. In her article she showed that, using an Atomic Force Microscope (AFM), one can successfully produce nano-patches of well oriented, surface-immobilized ssDNA-molecules with different oligonucleotide sequence, on a gold surface, in a nanoarray format, and use such nano-patches to immobilize via Watson-Crick base-pairing different protein-DNA conjugates (a methodology named DNA Directed Immobilization (DDI)). Such a DNA-barcoding protein nanoarray was then used to study biomolecular interactions. The authors monitored bio-recognition events exposing the device to the biosample and detecting the protein of interest measuring the change in the topographic height of each nano-patch with respect to an atomically flat gold surface. Based on AFM differential height measurements, Bano et al. demonstrated that, using a proper gold passivation, unspecific binding can be avoided, even when detection is carried out within a complex medium (serum, plasma). Moreover they proved to be able to detect at least 3 different biorecognition events simultaneously, with a detection limit of few tens of pM [8, 9]. Although expandable in their multiplexing capability (up to 10 different DNA sequences, and after that protein-DNA conjugates, can be grafted on the same surface, without damaging the sample), such protein nanoarrays are not easily integrable into a microfluidic platform. AFMs are in fact very expensive machines and, normally, must be operated by a trained technician in order to assure a correct usage and will hardly be employed in diagnostics during medical treatment. Faster and cheaper devices need to be implemented. In the last decades many setups for point of care medical diagnostics have been proposed. They can be roughly divided into three big classes according to the readout technique: optical, mechanical or electrical. The first class includes the well-established ELISA assay [10, 11, 12], other different fluorescence-based assays [7], surface plasmon resonance (SPR) [13, 14, 15] and light scattering/absorption assays [Raman [16, 17], FTIR [18, 19], light scattering [20]]. The second class relies on 4 1 Introduction the variation of mechanical properties induced by the bio-recognition event and include, among others, micro cantilevers and resonators [21, 22, 23] and acoustic-wave sensors [24, 14]. These first two classes are very appealing in terms of limit sensitivity and are well suited to laboratory applications; however, as mentioned for the AFM-based sensors, they are generally built into large instruments, whose cost cannot easily be decreased to make them viable for systematic point-of-care diagnostics. Conversely, detectors belonging to the third class, based on direct readout of an electrical signal, once miniaturized, could be easily integrated into portable electronics, at a fraction of the dimensions and total cost of competing instruments. Electrochemical chips integrated in electronic circuitry, combined with a microfluidic network that enables fast handling and reduces sample amount, were already proposed [25, 26] and are one example of the lab-on-a-chip concept [27, 28, 29, 30, 31, 32]. Within this family, capacitance-readout-based detectors represent one of the most promising strategies [24, 33, 34]. In our work we focused our attention towards this class of bio-sensors. We first explored the behavior of a two-electrode detector in a microfluidic channel (see Figure 1.1). The idea was to functionalize the gold electrodes with bio-probes, as in the work of Bano et al [8], and measure the electrical response of the device upon binding of a biological probe. Fig. 1.1. Representation of a two-electrode detector in a microfluidic channel. The potential is applied across the two electrodes which, on their own, are connected by the ionic solution present in the microchannel (thick black line that connects the inlet- and the outlet-tube). The interfaces electrode/electrolyte are the key-parts of the circuit, where there is a discontinuity in the nature of the charge carriers which, electrons in the lead-wires, become ions in solution. From Figure 1.1 we note that the potential is applied across the two electrodes and the channel assures the electrical connection. The key-parts of the circuit are the two electrode/electrolyte interfaces. At these two points in fact, there is a discontinuity in the nature of the charge carriers which, electrons in the leadwires, become ions in solution. How the passage from electrons to ions takes place is the subject of study of electrochemistry. Electrochemistry is not at the core of this thesis and thus we will not discuss it in details. At this point we limit ourselves to the consideration that, in order to allow charge-transport from 1 Introduction 5 the electrode to the electrolyte and vice versa, electrochemical reactions must take place. These reactions change the ionization state of the atoms at the surface and thus are not desired when building a detector for bio-molecules that shall not be changed by the detector itself. Nonetheless there is a way in order to avoid electrochemical reactions at the gold surface. The idea is very simple and consists in the application of an AC-voltage instead of a DC-voltage across the two electrodes. In this way the charge does not have to travel across the interface and the electrons in the wires and the ions in solution can simply oscillate around a central position. At the interface the moving ions in solution are periodically attracted and repelled from the surface and, in first approximation, behave like a conventional capacitor. This capacitance is called double layer capacitance, C DL , and is formed by the electrode and the layer of mobile ions in the liquid within a distance equal to the Debye length of the solution. This capacitance was proposed for the first time in 1853, when Helmholtz tried to model the electrode/electrolyte interface. His idea was simple but very intelligent. Knowing that, at equilibrium, the charge in the metallic electrode is distributed at its surface, he imagined that the ions in solution behave in the same way and distribute on a surface at a distance d from the electrode. The ions counterbalance exactly the charge on the metal and form, together with the electrode, the double layer capacitance, C DL . After this first model several scientists refined the theory but the name given by Helmoltz to the double layer capacitance remained (more details can be found in Appendix A). The distance within which the ions in solution interact with the charged surface of the electrode is called Debye-length and, for a symmetric monovalent salt, is defined as: 0 RT (1.1) 2F 2 c0 where R is the gas constant, T the absolute temperature, F the Faraday constant, c0 the molar conr κ −1 = centration of the salt in solution, and 0 the dielectric constant of liquid and vacuum, respectively. κ−1 bears information about the electrostatic screening effect of the solution and, as we can see in Equation 1.1, decreases for increasing ionic strength (more details can be found in Appendix A). With the knowledge that the electrode/electrolyte interface behaves like a capacitor, we can build the electrical equivalent model of the microfluidic system with two electrodes shown in Figure 1.1. In Figure 1.2 we present it. From Figure 1.2 we highlight the three main components of the equivalent circuit: C DL , C stray and Rchannel [33, 35]. As already explained, C DL models the interface electrode/electrolyte whereas C stray and Rchannel idealize the capacitance that exists across the two electrodes and the ionic resistance of the channel, respectively. In the model shown in Figure 1.2 the resistance of the wires was considered negligible and therefore it is not reported. In literature one finds many examples of bio-detectors based upon these three sensible parameters [2, 35, 36, 37]. In 2009 for instance, Vlassiouk et al. realized the prototype of a fluidic diode exploiting the phenomenon of surface conductance within polyethylene terephthalate (PET) nanochannels [38]. In their work the authors studied the resistive behavior of the system (Rchannel in Figure 1.2) and show its huge biosensing capabilities obtained by an appropriate choice of the functionalization of the channels. The device proposed by Vlassiouk et al. is one example of the detectors based on the measurement of the surface conductance within nanochannels [39, 39, 37]. This type of devices promises to have a great impact in the future of medical diagnostics but, at the same time, the fabrication of the nanochannels is still very complex and cannot guarantee the cost reduction and the multiplexing capabilities which 6 1 Introduction Fig. 1.2. Electrical model of the physical system shown in Figure 1.1. C DL models the interface electrode/electrolyte; C stray the capacitance that exists across the two electrodes while Rchannel represents the ionic resistance of the channel. we are seeking. Moreover in these devices the functionalization with biomolecules is not as easy as for gold surfaces. For all these reasons we decided to concentrate our attention on the detectors based on capacitance. These sensors work upon changes of the molecular layer at the electrodes and thus were the ideal candidates to be developed in the contest of the AFM measurements performed in our laboratory. In general, this type of measurements is classified as electrochemical impedance spectroscopy (EIS) and, according to the frequency of the applied voltage, the capacitance at the electrode/electrolyte interface or the stray capacitance between two adjacent electrodes is measured [2, 35]. At low frequencies (for our setup this means frequencies lower than 1kHz) the measurement is sensible to the electrode/electrolyte interface, at high frequencies (higher than 100kHz) to the stray capacitance. To measure the stray capacitance between two electrodes, interdigitated microelectrodes (IDEs) are normally preferred. For this kind of setup reference electrodes are not required, making the integration easier. IDEs, a scheme of their layout can be found in Figure 1.3, are normally produced via e-beam lithography [40, 41, 42, 43] and, in our opinion, are not the ideal candidates for detection of biomolecules. Measuring the stray capacitance across the electrodes in fact, is not the most sensitive way to capture the small variations occurring at the electrode/electrolyte interface connected to biorecognition events: a thickness variation of few nanometers at the interface, as for instance expected upon binding of a protein layer over an antibodies-covered surface, in order to be detected should induce a significant difference in the total capacitance of the detector which spans, according to the IDEs fingers interspace distance (d in Figure 1.3), over hundreds of nm. This variation is therefore very difficult to detect. Zou et al. [43] for instance, could not measure any impedance variation in the high frequency range ( f > 100 kHz) upon binding of mouse anti-rabbit IgG at a nanoIDE surface, despite the large concentration used, 250 µg/mL. Instead, they were able to measure impedance changes only at low frequencies, at which electrode/electrolyte interface capacitance dominates. Measuring at low frequency is thus preferable and became the focus of our work. In the past many authors tried to develop bio-detectors based on the capacitance at the electrode/electrolyte interface and we found out that they often noticed time instabilities and poor reproducibility [44, 45]. In order to limit these problems the authors devoted special effort to the quality of the elec- 1 Introduction 7 Fig. 1.3. Scheme of the interdigitated electrodes (IDEs). (a) Image of the two electrodes. The finger-like structures of the electrode on the left intersect the same structures of the electrode on the right. (b) Zoom-in of the IDEs fingers. Each finger has a width, w, and is separated from the next one by a distance, d. Both d and w can vary from few nanometers to some micrometers. trode surface. Yet in 1997, Mirsky et al. [45] studied the effect of length in alkylthiols self assembled monolayers (SAMs) on the capacitance signal: electrodes passivated with long thiols (15-16 methylene groups) were more stable in time than those passivated with short thiols (up to 10 methylene groups), which are known to be less densely packed. More recently, Carrara et al. performed atomic force microscopy (AFM) morphology measurements to address the short-time stability of the capacitance signal for a two-electrode setup as a function of the surface functionalization [44]. Although the authors claim that for more ordered SAMs no variations of the capacitance were observed within 10 min from the start of a measurement, they also state that a conditioning time of 24 hours was always necessary in order to obtain reproducible, stable measurements. Such a long waiting time is incompatible with a real-time point-of-care diagnostic tool. A possible solution to this problem is the implementation of a reference electrode. Nonetheless, complete electrochemical cells, consisting of working electrode (WE), counter electrode (CE) and reference electrode (RE), integrated into a microfluidic network, are yet difficult to realize, especially regarding RE miniaturization [46, 47, 48]. In the present experimental work we implemented and compared different biosensing platforms based on electrochemical impedance readout with the ultimate goal of performing label-free, real-time measurements of clinically relevant biomarkers. We functionalized microfabricated gold electrodes, and carried out capacitance measurements at low frequency, to highlight the small interface capacitance difference at the electrode/electrolyte interface occurring upon binding of relevant biomolecules. We addressed the issue of time stability and reproducibility of the sensor for two different configurations: a twomicroelectrodes, microfluidic device and a three-electrode electrochemical cell. In the latter, the working electrode and the counter electrode are in the micrometer scale while the reference electrode is a classical mm-sized Ag/AgCl electrode. We applied both configurations to the study of the hybridization of ssDNA SAMs on gold surfaces. The two-electrode setup turned out to suffer from a strong drift of the signal, which may last hours and makes any use of the device as a bio-molecular detector problematic. The three-electrode setup on the contrary, showed a very good stability in time and is, for this reason, a 8 Differential Capacitance good candidate for further development; in a preliminary test, it enabled us to measure, in real time, the hybridization kinetics of a ssDNA SAM. Differential Capacitance As already stated, in this work we focus our attention on the measurement of the capacitance at the electrode/electrolyte interface. Practically we will always speak of differential capacitance, Cd . In our configuration Cd is equal to C DL , the double layer capacitance already introduced in the previous section, and is defined as: C DL = Cd = ∂σ M ∂φ (1.2) where σ M is the charge density on the metal electrode and φ the potential difference between electrode and solution. In other words Cd measures the charge density change at the metal surface for a small variation of the applied potential. In the case of a bio-functionalized metal electrode immersed in a saline solution, Cd can be modeled with two capacitances [49, 50]: the capacitance due to the absorbed layer of molecules (Cmol in Figure 1.4) and the one due to the ions in solution (Cions in Figure 1.4). Fig. 1.4. Idealization of the electrode-electrolyte interface. The first layer, directly in contact with the gold surface, consists of the biological molecules and is modeled by the capacitance Cmol . The ions just above the first layer are modeled with a second capacitance, Cions , in series with the first one. Variations of Cmol arise upon molecular recognition. From Figure 1.4 we note that the two capacitances are connected in series and therefore the smaller of the two dominates according to the following relation, which describes the total capacitance, CT OT , as a function of the two capacitances. CT OT = CmolCions Cmol + Cions (1.3) Differential Capacitance 9 Normally, Cions is larger (densities of the order of 40µF/cm2 ) than Cmol and thus the latter dominates the measured value of Cd . Variations of Cmol arise upon molecular adsorption on the electrode surface and include height changes, substitution of water molecules in the biological layer, and changes in the electrical charge density. We note that the series of capacitances proposed above is the simplest approximation capable of modeling data. More accurate models, which include constant-phase elements and charge-transfer resistance have been proposed, but they are beyond the scope of this work [33]. The adopted procedure to measure Cd was tuned according to the electrochemical configuration in use. A detailed explanation of the experimental procedure can be found in Chapter 2. 2 Materials and Methods 2.1 Materials The used DNA- and alkanethiol-molecules were purchased from Sigma-Aldrich. They are sold in the form of lyophilised powder or solution and were used as received. Basic information and characteristics are listed in Table 2.1. molecule format molecular weight melting temperature HS-cF5 powder (g/mol) (°C) sequence 6861 65.4 5’-[ThiC6]-CTTATCGCTTTATGACCGGACC F5 powder 6812 65.4 5’-GGTCCGGTCATAAAGCGATAAG MCH solution 134.24 / HS (CH2 )6 OH TOEG6 solution 356.47 / CH3 O(CH2 CH2 O)6 CH2 CH2 S H 230.45 / HS (CH2 )1 3CH3 C14 solution Table 2.1. General features of the purchased DNA- and thiol-molecules. The melting temperature is referred to the dsDNA. The ssDNA that binds to the gold surface is functionalized at the 5’-end with a thiol composed by a chain of 6 carbon-atoms. The sequence is referred as the sequence of the nucleotides for the DNA-molecules and the sequence of the atoms for the thiols and the top terminated oligoethylene glycol (TOEG6). For the experiments we prepared aliquots of the DNA solutions (DNA concentration c = 100 µM ) by dissolving the received amount of powder in a solution of T E NaCl 1 M at pH = 8. T E is the acronym of the buffer solution composed by T ris 10 mM plus EDT A 1 mM. Stock solutions of the different salts were prepared weighting the right amount of powder on a balance (accuracy 1 mg) and adding milli-Q water (resistivity ρ = 18.2 MΩcm) by means of calibrated pipettes. The other concentrations were prepared diluting the stock solutions. Prior to use all saline solutions were filtered using filters from Millipore with pores size, d = 0.22 µm. Some important characteristics of the used salts are listed in Table 2.2. In the same table we list some features of Tris, an organic compound used in buffer solutions, and EDTA, a chelating agent for multivalent metal ions like Ca2+ and Fe3+ . In our work we used this two molecules in order to produce the TE-buffer, the solution used to dilute the DNA-molecules. 12 2 Materials and Methods Company Type molecular weight (g/mol) purity NaCl Sigma Aldrich powder 58.44 ≥ 99% KCl Sigma Aldrich powder 74.55 ≥ 99% PBS Sigma Aldrich powder / / KH2 PO4 Sigma Aldrich powder 136.09 = 99% Tris Sigma Aldrich powder 121.14 ≥ 99.8% EDTA Sigma Aldrich powder 292.24 > 99% Table 2.2. General features of the used salt. Stock solutions of the different salts were prepared weighting the right amount of powder on a balance (accuracy 1 mg) and adding milli-Q water (resistivity ρ = 18.2 MΩcm) by means of calibrated pipettes. PBS has no specific molecular weight because it is a mixture of different salt (NaCl 137 mM, KCl 2.7 mM, Na2 HPO4 10 mM, KH2 PO4 2.0 mM). Besides salts and biological molecules, during the Ph.D. we made extensively use of the organosilicon compound Polydimethilsiloxane (PDMS). PDMS has been used in the fabrication of the microchannels for the two-electrode setup. In the next section we explain how we employed PDMS in the soft lithographic process. The main characteristics of the used PDMS are listed in Table 2.3. Producer Dow Corning product name Sylgard 184 contact angle ≈ 110 deg (hydrophobic) mix ratio 10:1 base to catalyst volume resistivity 1.2 · 1014 Ωcm Cure time @ 110 °C(hot plate) several minutes Table 2.3. Main features of the used silicon compound: PDMS. The data have been copied from the Dow Corning website. 2.2 Device Fabrication 13 2.2 Device Fabrication The fabrication processes we employed for the realization of the micro-electrodes and the microfluidicchannels are photo- and soft-lithography, respectively. In Figure 2.1 we show, in a schematic manner, all the fabrication steps one has to go through during photolithography. First of all the clean slides are dehydrated at 200 °C for 5 min in order to increase the adhesion of the resist (a). Successively the slides are rapidly cooled down in a nitrogen stream and immediately spin-coated with a UV-photo-sensible resist (b). According to the fabrication needs, one can vary the film thickness by varying the rotational speed of the spin-coater. After spinning, the slides are put on a hot plate in order to perform the pre-bake of the resist (c). Temperature and baking time are defined by the resist and its thickness. At this point the samples are exposed to UV-light through a Cromium mask and the mask pattern is transferred into the photo-sensible film (d). The type of the resist, positive or negative, defines how the mask image is transferred. For positive resist the exposed areas are removed during development whereas, for negative resists, the exposed areas remain on the slides after development. From Figure 2.1 we see that after light exposure the process varies a little whether we use a positive or a negative resist. In the first case the samples are immediately developed whereas, in the second one, a post-bake process must be performed. In this thesis-work we employed mostly S1818 and SU8 as a positive and negative resit, respectively. Note that the steps described in Figure 2.1 may change a little as a function of the specific resist. In Appendix B we list all the details regarding the resists and the experimental procedures that we employed. In Appendix B one can further find the description of all the instruments used in the photo-lithographic process. In order to fabricate the micro-electrodes, after the described photolitographic steps, one has to perform the metallization of the glass slides in an evaporator. In Figure 2.2 the procedure we adopted is described. First the micro-patterned microscope slides (S1818 is used as positive resist) are inserted in an e-beam evaporator and two metal layers, 50 nm of Titanium (adhesion layer) and 50 nm of Gold, are deposited (a and b) under high-vacuum. Following evaporation the slides undergo lift-off (c) and the sacrificial resist is removed. In this process the samples are immersed in an acetone bath overnight and eventually rinsed thoroughly with acetone and isopropanol. After lift-off the sample with the twoelectrode configuration can be immediately used. The ones with the three-electrode configuration instead, undergo another complete lithographic sequence in order to define the area of the working and counter electrode in contact with the solution and isolate the rest. in this last part of the electrodes fabrication we use S1818 as positive resist and we perform an aligned lithography using the mask aligner MJB3 by Karl Suss, Germany. A picture and a description of this instrument can be found in Appendix B. Examples of the slides with the micro-electrodes used in the two- and three-electrode configuration, are shown in Figure 2.3 and Figure 2.4, respectively. The microfabricated electrodes of the two-electrode setup are composed by two gold pads for wire-connection and two gold paths that act as electrodes in the complete setup. The paths have a width of 100 µm and their separation varies from 1000 µm to 300 µm. The WE in the three-electrode setup has a diameter, d = 100 µm, and is connected to the gold pad by a path with variable width. The RE is an arc with a smaller diameter of 300 µm that encloses the WE. The path that connect WE and RE with the connection-pads are isolated from solution by a layer of S1818 with a thickness of 2.5 µm. 14 2 Materials and Methods Fig. 2.1. Fabrication steps during photolithography as a function of the used resist (positive or negative). After dehydration (a), the slides are spin-coated with a photosensible resist (b) and baked on a hot plate (c). The slides are then ready for UV-exposure (d) and then, as a function of the resist, for post-bake (only negative resists) and development (e) and (f). In order to fabricate the microfluidic-channels we employed soft lithography. This technique is a molding process and can produce many replicas of the same model using a curable elastomer. In our work we used Polydimethylsiloxane (PDMS) as elestomer and SU8 as negative resist in order to fabricate the mold of the channel. The process is very simple and can be described with only three steps (Figure 2.5). First the operator pours the elastomer on top of the model and waits until the PDMS has uniformily distributed (a). Second the mold is transferred on a hot plate and the PDMS cured for several minutes at 110 °C (b). Third, the solidified elastomer is delicately peeled off the stamp (c). We note here that the PDMS is a two-component elastomeric solution: PDMS and cross linking solution in a ratio of 10 : 1. In Section 2.1 one can find all the details of the used PDMS. The final dimensions of the produced channels are: width, w = 100 µm, height, h = 67 µm, length, l = 3 cm. In order to connect the channels with the fluidic pump-system we drilled holes with a diameter, d = 1/16 inch, at the two ends of the channels and we inserted peek outlet and inlet tubes. In Figure 2.6 we show an example of the PDMS block with the peek orange tubes and the microfluidic channel. 2.2 Device Fabrication 15 Fig. 2.2. Electrodes fabrication. After lithography the microscope slides are inserted in an e-beam evaporator and the metals, Ti as adhesion layer and Au, are evaporated (a,b). After metallization the metals in excess is eliminated via lift-off (c). Fig. 2.3. Picture of the microfabricated electrodes for the two-electrode setup. The gold electrodes were fabricated on half a microscope slide and are composed by two thin metallic films: 50 nm of gold and 50 nm of titanium. Ti acts as adhesion layer between gold and glass. 16 2 Materials and Methods Fig. 2.4. a) Picture of the microfabricated WE and CE for the three-electrode setup. The gold electrodes were fabricated on half a microscope slide and appear black in the image. The patterned insulation layer (S1818; thickness: 2.5µm) appears pink. b) Zoom-in of the central part of the picture. The diameter of the WE in contact with the solution is 100 µm. The patterned resist used to electrically insulate the electrodes is clearly recognizable as the darker-gray outer area of the image. Fig. 2.5. Fabrication steps during soft-lithography. First the PDMS is poured on the mold of SU8-100 (a) and then transferred on a hot plate for curing (b). Finally the PDMS is gently peeled-off the slides (c). 2.2 Device Fabrication 17 Fig. 2.6. PDMS block with peek orange tubes and the microfluidic channel. The channel connects the inlet and outlet tubes and is visible as a light grey line among them. 18 2 Materials and Methods 2.3 Experimental Setup In order to connect the micro-electrodes with the electrical instruments and thus perform the measurement, we developed two different sample holders according to the experimental setup. The sample holders for the two-electrode and the three-electrode devices are shown in Figure 2.7 and 2.8, respectively. From the pictures we note that in both cases the slides with the micro-electrodes are costrained between two plexiglas slides. In the first case the micro-channel is placed orthogonally to the electrodes exposing to the solution an area of the electrode of about 100x100 µm2 . In the second case, because of the dimension of the reference electrode (diameter, d = 5 mm), the measurements were not carried out in the microfluidic channel. Instead we used a small pool with a diameter of 6 mm and a height of 4 mm, holding a 100 µL volume. In this way the reference Ag/AgCl pellet electrode can be inserted directly in the solution through the hole in the top plexiglas slide and placed just above the microelectrodes (see Figure 2.8a). The electrical connections were developed in two different stages of the project and are a bit different. In the case of the two-electrode setup, connection wires are directly soldered at the two gold pads (5x5 mm2 ) with an Indium drop and then connected with the instrument. In the three-electrode setup on the contrary, the electrical signal is collected from the gold pads (4x4 mm2 ) by a circuit board of custom design with SMA connectors and spring-loaded pins (see Figure 2.8b). In order to shield the device from electromagnetic interferences we further designed and realized a very robust Faraday cage (see Figure 2.9). The cage is made of Aluminium plates with a thickness of 1cm and has a total weight of roughly 10kg. In this way also mechanical vibrations can be effectively shielded and measurements at low frequencies become easier. Fig. 2.7. Sample holder for the two-electrode devices. The microscope slides with the micro-electrodes are costrained between two plexiglas slides: the micro-channel is placed orthogonally to the electrodes exposing to the solution an area of the electrodes of about 100x100 µm2 . Connection wires are directly soldered at the two gold pads (5x5 mm2 ) with an Indium drop and then connected with the instrument 2.3 Experimental Setup 19 Fig. 2.8. a) Sample holder for the three-electrode devices. The microscope slides with the micro-electrodes are costrained between two plexiglas slides. Because of the dimension of the reference electrode (diameter, d = 5 mm), the measurements were not carried out in the microfluidic channel but instead we used a small pool with a diameter of 6 mm and a height of 4 mm, holding a 100 µL volume. The reference Ag/AgCl pellet electrode is inserted directly in the solution through the hole in the top plexiglas slide and placed just above the microelectrodes. b) The electrical signal is collected from the gold pads (4x4 mm2 ) by a circuit board of custom design with SMA connectors and spring-loaded pins. Fig. 2.9. a) Faraday cage designed in order to shield the external electromagnetic noise. The cage is made of Aluminium plates with a thickness of 1 cm and has a total weight of roughly 10 kg. The weight prevents mechanical vibrations at low frequencies to disturb the measurement. b) The Faraday cage permit the use of 5 BNC connection cables. The BNC connector are isolated from the cage but can be grounded through a wire (green wire in the picture). The cage itself can be grounded through a banana-connector. 20 2 Materials and Methods 2.4 Instruments: Electrical and Topographic Characterization 2.4.1 DSO Lock-In Amplifier: Model SR830 Measurements with the two-electrode setup were performed with the lock-in amplifier, model SR830 by Stanford Research Systems. An image of the instrument is shown in Figure 2.10 while its main characteristics are summerized in Table 2.4. The main advantage of the lock-in amplifier is its capability to filter out all the electrical noise with a frequency different than the measuring one. In other words, the lock-in amplifiers can amplify very low voltage- and current-signals at a certain frequency and filter out all the noise coming from the other frequencies. Readers interested in the technical details can find a very nice and short explanation in the section <<What does a Lock-In measure?>> in the manual of the instrument. The measurements we performed with the lock-in are frequency scans. An AC voltage with frequency ranging from 10 Hz to 90 kHz and a root mean square (rms) amplitude of 10 mV was applied across the electrodes and the current flowing in the channel was amplified with a transresistance amplifier, Femto DLPCA 200 (see Figure 2.12), and measured. In order to automatize the scans we wrote a procedure using LabView, a software powered by National Instruments. In Figure 2.11 we schematize all the steps implemented by the program in order to complete the measurements. Model SR830 Producer Stanford Research Instruments Operation mode automatic and programmable via LabView Output frequency range 1 mHz-102 kHz Output voltage range Vrms 4 mV-5 V (error 1%) Output impedance 50 Ω Voltage input impedance 10 MΩ + 25 pF in parallel Current input impedance 1 kΩ to virtual ground Voltage Inputs single-ended or differential Table 2.4. Main features of the lock-in amplifier, SR830 by SRS. Fig. 2.10. Front panel of the lock-in amplifier, model SR830 by Stanford Research Systems. 2.4 Instruments: Electrical and Topographic Characterization 21 Fig. 2.11. Steps implemented by the program written for the lock-in amplifier. After the initialization of the parameters the programs enter the main loop and leave it only when the set frequency is higher than fEND , the end frequency of the measurement. Fig. 2.12. Picture of the transresistance amplifier DLPCA 200 by Femto. The current that enters is amplified and transformed in a proportional voltage signal. The amplifier can be used both for DC and AC current-signal and the amplification factor can be tuned in a very broad range, from 103 to 1011 V/A. According to the amplification factor the band width of the instrument decreases from 500 kHz to 1 kHz. 22 2 Materials and Methods 2.4.2 Heka Bipotentiostat PG 340 usb Measurements with the three-electrode setup were performed with the bipotentiostat, model PG340 usb by Heka. An image of the instrument is shown in Figure 2.13 while its main characteristics are summarized in Table 2.5. The advantage of the bipotentiostat, when compared with the lock-in amplifier, is the presence of a third reference electrode, which, due to the high stability of its potential in contact with an electrolitic solution, assures a fine control of the absolute value of the applied potential at the working electrode. The performed measurements are essentially the same as with the lock-in. An AC voltage with amplitude of 10 mV at different frequencies (between 100 Hz and 400 Hz) was applied across WE and RE and the current flowing between WE and CE amplified and measured by the instrument (see Figure 2.11). At each frequency we collected 200 complete periods from which we computed the root mean squared value of the measured current, Irms , and the relative uncertainties using error propagation analysis. Eventually we fitted the Irms value with a linear fit in frequency and we computed the value of the capacitacne at the electrode/electrolyte interface. In Figure 2.14 we show the scheme of the procedure that we wrote in order to analyze the experimental data. The procedure was implemented in Igor, a software for data analysis developed by Wavemetrics. The procedure was inserted in a loop in order to analyze the variations of Cd as a function of the time. Model PG340 usb Producer Heka Operation mode automatic and programmable via Potmaster Output frequency range DC-20 kHz Compliance Voltage ± 20 V Output Current ±1A Input impedance 100 GΩ + 1.5 pF in parallel Measurable current range 1 nA-1 A Output channels Disk- and Ring-potentiostat setup possibilities 3-electrodes and 4-electrodes Table 2.5. Main features of the bipotentiostat, PG340 usb by Heka. 2.4 Instruments: Electrical and Topographic Characterization 23 Fig. 2.13. Front panel of the bipotentiostat, model PG340 usb by Heka. Fig. 2.14. Steps implemented by the procedure written in order to analyze the experimental data collected by the bipotentiostat. After data acquisition the procedure computes the Irms value for each measured frequency. Afterwards the program plots the Irms -values as a function of the frequency and fits them linearly. From the fit the value of Cd can be calculated. This sequence can be repeated for every complete data set as, for example, when one wants to measure variations of Cd in time. 24 2 Materials and Methods 2.4.3 Atomic Force Microscope: MFP 3D Topographic measurements were conducted on the three-electrode devices and were performed with the atomic force microscope, model MFP 3D by Asylum Research, operating in contact mode. An image of the instrument is shown in Figure 2.15 while its main characteristics are summarized in Table 2.6. The measurements performed with the AFM are presented in Chapter 4 and consist of shaving experiments of the DNA-SAM. More details are given in the relative section. Fig. 2.15. Atomic force microscope: model MFP 3D by Asylum Research. Model MFP 3D Producer Asylum Research Operation mode contact and non contact X & Y Scan axes 90 µm Z scan axis 15 µm X & Y sensor noise 0.5 nm Z sensor noise 0.25 nm Noise of the optical lever < 0.02 nm in the bandwidth 0.1Hz − 1kHz Stage micrometer driven stage for mechanical alignment of cantilever tip and sample Table 2.6. Main features of the atomic force microscope, MFP 3D by Asylum Research. More information can be found on the Asylum Research web-page. 2.5 Experimental Procedures: SAM Formation and Hybridization Procedure 25 2.5 Experimental Procedures: SAM Formation and Hybridization Procedure DNA functionalization of the gold electrodes was carried out using the well-established procedure for DNA self assembled monolayers (SAMs) on gold [51, 52, 53]. Initially the electrodes (both in the twoand the three-electrode setup) are wetted with a drop of a high-ionic-strength buffer, T E NaCl 1 M, containing thiolated (C6) ssDNA 1µM (22 bases long). The density of the SAM can be controlled varying the immersion time. Principally we used two different immersion times: low density SAM were realized keeping the samples in contact with the functionalizing sulution for 10 min whereas high density SAM were formed increasing the time to 2 h. According to Georgiadis et al. the obtained SAM densities on the gold surface using these parameters are: 2−3x1012 molecules/cm2 for LD SAM and 1x1013 molecules/cm2 for HD SAM [51]. After the DNA-SAM formation, in order to remove unspecifically bound DNA-molecules, the electrodes were left for 1 h in contact with a solution of T E NaCl 1 M containing mercaptohexanol (MCH) 1 mM. MCH-molecules compete with unspecifically bound ssDNA-molecules (i.e. physisorbed through the backbone-gold electrostatic interactions), removing them and chemisorbing onto the surface through S-Au bond (≈ 2 eV). An idealization of the mix SAM ssDNA + MCH is shown in Figure 2.16. Fig. 2.16. Cross section of the mixed SAM formed by ssDNA- and MCH-molecules. MCH-molecules compete with unspecifically bound ssDNA-molecules, removing them and chemisorbing onto the surface through S-Au bond (≈ 2 eV). Finally two-electrode devices were thoroughly rinsed with milliQ water (resistivity 18.2 MΩcm) and dried under a nitrogen stream. Three-electrode devices instead, were not dried but rinsed directly with the buffer solution used for the measurements, KCl 10 or 100 mM. The hybridization step was performed differently for the two configurations. Two-electrode-devices were fluxed with a solution of T E NaCl 1 M containing the complementary ssDNA strand 1 µM at a rate of 0.02 mL/h for 1.5 h. Afterwards, the measuring solution, KCl 10 mM, was fluxed for 1 h in order to rinse the microchannel and establish the same measuring conditions as for ssDNA. Three-electrode-devices were wetted with a drop of the hybridizing solution, T E NaCl 1 M containing the complementary ssDNA 1 µM, kept in contact for 1.5 h, after which the devices were thoroughly rinsed with KCl 10 or 100 mM immediately before the measurement. In the case of real-time measurements of the DNA-hybridization kinetics, the hybridization was performed with 1 nM complementary ssDNA in KCl 100 mM buffer solution. SAM of thiols, e.g., tetradecanethiol (C14) and mercaptohexanol (MCH), were produced as well. We formed them by immersion of the gold electrodes in the solution containing the thiols overnight. The 26 2 Materials and Methods solution varied as a function of the molecules. We used ethanol for the alkanethiols and aqueous buffer for MCH. From literature we know that thiols on gold form tightly packed monolayers, in which the sulfur atoms bind covalently on gold and the alkyl chains interact via van der Waals’ interaction [54] (they form hexagonal close-packed structure where the next neighbor distance is d = 0.5 nm). Differently from DNASAM, which interact via electrostatic forces due to the the negative charges of the DNA-backbone, SAM composed by thiols passivate very well the gold surface due to the higher packing density (up to 4 orders of magnitude higher than DNA-SAM). 3 Two-Electrode Configuration In this chapter we start the presentation of the experimental results obtained during the Ph.D. This part is the first of the two chapters describing the results obtained with the two-electrode configuration and the ones obtained with the three-electrode configuration. The division reflects the development and the progressive sophistication of the device. During the Ph.D. in fact, we continuously tried to improve the detector in terms of stability and reproducibility and the last results are the outcome of this effort. In the following pages we present the experiments we performed with the two-electrode configuration. We describe thoroughly the electrical characterization of the system, we highlight the limitations of the setup and present how we decided to proceed in order to overcome those limitations. This process will naturally lead to the three-electrode configuration described in the last part of the thesis. 3.1 Electrical Characterization As explained in 2.4, in order to perform an electrical characterization of the two-electrode setup, we applied an AC-voltage with frequency ranging from 10 Hz to 90 kHz and a root mean square (rms) amplitude of 10 mV across the electrodes. The ionic current flowing in the channel was then measured with the lock-in amplifier. In order to understand the general behavior of the system we first consider the Bode-plot shown in Figure 3.1. In Figure 3.1 we plot on the same graph the root mean squared current, Irms , and the phase delay of the signal with respect to the applied voltage, φ, as a function of the applied frequency, f . This graph was obtained for a solution of KCl 1 mM. Immediately we can notice 3 different regions. In the first one, frequency range 10 Hz to 100 Hz, Irms increases linearly whereas the phase is always above 40 degrees. In the intermediate one, between 400 Hz and 20 kHz, Irms becomes almost independent of f and the phase does not exceed 20 degrees whereas, for frequencies higher than 50 kHz, the system recovers the initial behavior. We note here that the decrease of φ above 50kHz is an artifact of the measurement induced by the cut-off frequency of the current/voltage amplifier and therefore we will not consider it in the discussion. As already mentioned in the introduction, we can explain this behavior using the electrical model shown in Figure 1.2. According to that model, the electrode/electrolyte interface can be modeled by a capacitance, Cd , the fluidic channel by an ionic resistance, Rchannel , and the interaction which exists between the two electrodes by another capacitance, C stray . Therefore, depending on the frequency of the applied 28 3 Two-Electrode Configuration Fig. 3.1. Root mean squared current, Irms , and its phase, φ, as a function of the applied frequency, f , for a solution of KCl 1 mM and an applied potential, Vrms = 100 mV. The current is shown as red circles, the phase as green squares whereas the blue dashed lines represent the fits obtained using equations 3.1 and 3.2. voltage, different parts of the circuit prevail in determining Irms and φ. In our setup, since Cd is much bigger than C stray (1 nF vs 1 pF), Cd dominates at low frequencies, whereas C stray contributes mostly at high frequencies. In the intermediate frequency regime the current is independent of frequency, and is dominated by Rchannel . A plateau is therefore measured, whose value depends on the ionic resistance of the solution in the fluidic network. According to the model shown in Figure 1.2 we can further derive the theoretical shape of such a circuit. The Equations, which describe Irms and φ as a function of the angular frequency, ω = 2π f , read: Irms q V0 ω ω2 R2Cd4 + (Cd + C stray + ω2 R2Cd2C stray )2 = √ 2(1 + ω2 R2Cd2 ) φ = arctan Cd + C stray + ω2 R2Cd2C stray ωRCd2 (3.1) (3.2) With Equations 3.1 and 3.2 we can fit the experimental data and study in what extent the model approximates the behavior of the two-electrode setup. The fits we obtained are represented in Figure 3.1 by the blue dashed curves. The current was fitted for all the experimental data-points whereas the phase only from 10 Hz to 40 kHz. From the graph we note that the model fits reasonably well the behavior of the system above 100 Hz while it diverges below this threshold. This discrepancy however does not surprise us. The proposed model of the electrode/electrolyte interface in fact, is only an approximation and obviously cannot completely describe its real behavior. Nevertheless the proposed model is surely 3.1 Electrical Characterization 29 valuable since it catches the main features of the two-electrode system. Moreover we would like to note that the higher the ionic strength, the better the capacitance approximation of the interface models the experimental data. In Figure 3.2 we show an example for KCl 10 mM. In this graph it becomes clear that the model, at this ionic concentration, fits reasonably well Irms throughout the entire frequency range. The reason is that the ions in the diffusive layer (see Figure 1.4 and Appendix A) are confined within a distance from the surface given by the Debye length, κ−1 (Equation 1.1). The smaller the Debye length, the better the capacitor approximation of two planes of charges separated by a distance κ−1 , introduced in the first chapter of the thesis, can model the electrode/electrolyte interface. Fig. 3.2. Root mean squared current, Irms , and its phase, φ, as a function of the applied frequency, f , for a solution of KCl 10 mM and an applied potential, Vrms = 100 mV. The current is shown as red circles, the phase as green squares whereas the blue dashed lines represent the fits obtained using equations 3.1 and 3.2. At this point we performed measurements as a function of the ionic strength of the solution in order to fix the proper salt concentration to better highlight differences in the differential capacitance at the electrode/electrolyte interface. In Figure 3.3 we show the impedance of the system, Z = Vrms /Irms , as a function of the frequency, f , for KCl at 6 different concentrations, from 1 µM to 100 mM. The profiles shown in Figure 3.3 correspond to increasing salt concentration from the top red to the bottom purple curve. Going from one profile to the next one the salt concentration increases by one order of magnitude. From the analysis of Figure 3.3, we see that the contribution of Cd to the measured impedance is highlighted using either KCl 10 or 100 mM (orange thick line and purple dashed line in Figure 3.3). Eventually we decided to use KCl 10 mM, concentration at which we can simultaneously extract information on Cd and Rchannel . As we can see from the orange curve in Figure 3.3 in fact, Cd dominates the 30 3 Two-Electrode Configuration Fig. 3.3. Impedance, Z, vs frequency curves for KCl at 6 different concentrations, from 1 µM (red dashed curve) to 100mM (purple dashed line). The salt concentration increases for every profile by one order of magnitude going down from the red curve at the top of the graph. The orange solid line represents the Z-values for a KCl concentration of 10 mM and is the solution of choice for the rest of the experiments in this chapter. impedance behavior of our system up to 1kHz while, from fitting the region from 1 kHz to 100 kHz, the contribution of Rchannel can be deduced. The possibility to measure Rchannel and Cd simultaneously is of crucial importance to calibrate the system: the same Rchannel in fact, guarantees that the performed measurements correspond to the same ionic strength in the channel. Since at 100 mM we cannot monitor Rchannel , we selected KCl 10 mM for our successive experiments. We note here that biological environments (e.g. extracellular environment) present higher ionic strength, several hundreds of mM, and thus a KCl concentration of 100 mM would have been preferable. In this phase of device-prototyping however, we preferred to have a better control of the measurements. Once we will be sure that the measurements are reproducible, we will change the ionic buffer in order to test our device at conditions more similar to the biological ones. 3.2 DNA-Hybridization 31 3.2 DNA-Hybridization The first experiment we performed in order to test the capabilities of the two-electrode setup was the detection of the DNA-hybridization on the electrodes surface. DNA-hybridization is one of the most studied processes in biology [52, 31, 55]. It is at the base of life in cellular replication and, with the introduction of the DNA microchips [56], it has become the basis for the development of fast and reliable tools for genes analysis. In our setup we used DNA-hybridization in order to test the two-electrode device. First, as explained in 2.5, we functionalized the electrodes with a low density SAM of DNA-molecules and, successively, we collected several current vs frequency profiles (Irms vs f ). Afterward we fluxed in the microchannel a solution containing the complementary DNA-strands and we repeated the measurement (see 2.5 for further details on the experimental procedure). The experimental results are summarized, in a Log-Log scale, in Figure 3.4. Fig. 3.4. I vs f profiles as a function of DNA-hybridization (average of five independent measurements, in a 10mM KCl solution). Red solid line: electrodes functionalized with a SAM of ssDNA + MCH; blue dashed line: same electrodes after hybridization. In the high frequency range the profiles overlap well, indicating that the ionic strength of the solution is the same. Changes in the low frequency range indicate modifications at the electrode-electrolyte interface. Inset: zoom-in of the low frequency range; markers: experimental points, dashed lines: best-linear-fit, same color scheme as main plot. The measurements reported in Figure 3.4 are the result of the averaging of 5 different I vs f profiles recorded in a solution of KCl 10 mM and with an applied voltage, Vrms = 0.1 V. The 2 regimes that characterize the device are clearly visible. At high frequencies the device is almost completely resistive and the current is independent of the frequency. As we can appreciate from Figure 3.4 the two curves 32 3 Two-Electrode Configuration in this regime overlap very nicely. This is exactly what we expected since, in this region, the leading parameters are the ionic strength of the solution and the dimensions of the channel. Since the PDMS channel does not change from one measurement to the other, the overlap of the curves assures that the ionic strength of the solution is exactly the same upon refilling the channel, before and after hybridization (KCl 10 mM). At low frequencies, instead, the impedance is dominated by the differential capacitance at the electrode. In this regime the current increases linearly with f . To extract the corresponding capacitance values, we fit these curves with the following linear function: Irms = m f = VrmsCT OT 2π f (3.3) where m is the linear coefficient, Vrms the root mean squared amplitude of the AC applied potential, f its frequency and CT OT the total capacitance of the device. We decided to use a simple linear fit instead of Equation 3.1 since the latter includes the contribution of C stray . This part of the electrical model cannot be explored because of our experimental parameters, KCl 10 mM measuring solution and frequency range limited to f = 100 kHz, and thus the fitting procedure using Equation 3.1 was not as reliable as the one with the simple linear function. Since CT OT is the series of the two Cd s (see circuit model in Figure 1.2), we can compute Cd at the gold electrode as: m (3.4) Vrms π In the inset of Figure 3.4 we show the plot of the current as a function of the frequency restricted Cd = to the low frequency range together with the relative fits obtained by using Equation 3.3. The extracted average Cd values are (0.652 ± 0.002)nF and (0.559 ± 0.003)nF for ssDNA and dsDNA, respectively. The relative standard deviation was calculated over 5 independent measurements. From these data we compute a decrease in capacitance of ca. 14% upon DNA hybridization. This value is in agreement with the existing literature [57, 58, 35, 50] and can be explained by the height increase of the SAM upon hybridization, due to the different persistent length of ss and ds DNA (1 vs 50 nm [52, 59]), and by the replacement of water molecules (high dielectric constant, ) with DNA molecules (lower ) upon DNA pairing. Furthermore we note that the capacitance density at the electrodes measured with our setup is ≈ 5 µF/cm2 . This value is in agreement with literature [60]. 3.3 Time Behavior of the Two-Electrode Setup 33 3.3 Time Behavior of the Two-Electrode Setup After having successfully detected DNA-hybridization, we moved forward and tested the time stability of the two-electrode setup. After a careful study of the literature in fact, we found that two-electrode devices show a strong time dependence [44, 45]. First of all we checked the time behavior of Cd for clean, non-functionalized electrodes. In Figure 3.5 we plot the measured Cd over a time interval of 15 h. Fig. 3.5. Time behavior of Cd for clean, non-functionalized electrodes. Cd seems to reach stability only after a time intervall of 15 h and a percentage change of roughly 25%. From Figure 3.5 we notice the strong time decay of Cd , which, at the end of the measurement, amounts to roughly 30%. This strong time dependence raises several questions regarding the reliability of the electrochemical assay with the two-electrode setup and the need to check the value of Cd vs time. An absolute comparison of the values of Cd before and after a biological recognition event in fact, can also depend on the experimental procedure, i.e., on the time at which we carry out the measurement. For this reason we started to search for a possible solution of the time-drift and, according to several examples present in literature, we first tested different electrodes functionalizations. According to previous works in fact, passivation of the electrodes is a simple and effective way in order to reduce and eventually suppress the time decay shown by the two-electrode devices [44, 45]. As observed by Carrara et al. [44, 61] in fact, the capacitance signal can be influenced by the buffer ions in solution that, once at the electrode, can discharge onto it. A compact SAM can hinder this phenomenon and increase the time stability of the measured Cd . First of all we tested two different SAMS over time:C14 and ssDNA + MCH. In Figure 3.6 we plot Cd values obtained for these two different functionalization-layers on the electrodes plus the curve already presented in Figure 3.5 for the bare electrodes. Each profile has been normalized to its value at 34 3 Two-Electrode Configuration t = 0 in order to facilitate the comparison between them. It is well-known in fact, that stochastic variations from one device to the other make an absolute comparison unreliable [35]. Fig. 3.6. Relative capacitance change as a function of time for a control measurement (clean electrodes) and two different functionalizations using the two-electrode configuration. Red: bare electrodes, blue: electrodes functionalized with C14 thiols, green: electrodes functionalized with (ssDNA + MCH)-SAM. From the correspondent curves in Figure 3.6 (red, blue, green) we notice a quite pronounced variation over time of the capacitance. Within 9 hours, Cd changed by a minimum of 10 % up to a maximum of 25 % (bare electrodes). The functionalization of the electrodes leads to a better time stability but cannot completely solve the problem. A possible explanation could be the bad quality of the formed SAM. In the works already cited in fact [44, 61], Carrara et al. notice that a non-homogeneous SAM can promote formation of nanogrooves, through which buffer ions can penetrate into the film and discharge onto the electrode. In our case the decrease of capacitance versus time could be connected to a worsening of the SAM morphology due to electrochemically-induced partial desorption of thiolated molecules [62, 63, 64]. We believe however, that this process is unlikely because the AC potential applied to the electrodes (10 mV) is nominally much lower than the threshold required for electrochemical-induced molecular desorption (hundreds of mV). Therefore, even if in the two-electrode setup the potential of the electrode with respect to the ionic solution cannot be controlled precisely, it can hardly exceed the threshold value for thiol-desorption. A much more realistic explanation is that, in this setup, the potential at the electrodes with respect to the solution, which is not controlled by a reference electrode measurement, undergoes a drift during time. As a consequence, since Cd depends on the potential at the electrode [65] (Chapter 13), its value varies in time. In this second and, we believe, more realistic hypothesis, variations of Cd 3.3 Time Behavior of the Two-Electrode Setup 35 are intrinsic to the measurement setup and not connected to a worsening of the interface layer at the electrode. In order to completely overcome this problem, always present in the systems investigated so far, we decided to evolve our experimental setup into a three-electrode configuration (see Chapter 4). Nevertheless, before doing it, we further stressed the two-electrode devices and check in what extent they can be used for biodetection. In terms of integrability and ease of fabrication (see Introduction), the advantages of a two- over a three-electrode configuration in fact, are worth a more detailed study. Towards this goal we evaluated absolute variations of Cd as a function of the functionalization layer of the electrodes. In these experiments we were no more interested in the time behavior of the capacitance signal but instead in the absolute variations as a function of the SAM layer. For this reason we sorted out three fabricated devices with a very similar initial value of Cd measured in KCl 10 mM and we functionalized the electrodes with three different SAMs: Mercaptohexanol (MCH); ssDNA without MCH and ssDNA with MCH. After the functionalization we measured the value of Cd in time and we compared the results as a function of the SAM-layer. in Figure 3.7 we show the obtained profiles over a time period of 10 h. Fig. 3.7. Absolute capacitance change as a function of time for three different functionalizations using the twoelectrode configuration. Red: electrodes functionalized with a MCH-SAM, green: electrodes functionalized with a (ssDNA + MCH)-SAM, blue: electrodes functionalized with a ssDNA-SAM. On the right of the graph we depicted the situation in three different cartoon. (a) cross section of the SAM formed by the MCH-molecules alone (The alkyl chain is depicted as a zig-zag line, the S-atom and the OH-group by a blue and an orange circle, respectively). (b) SAM composed by the mix ssDNA- + MCH-molecules. (c) SAM composed by the DNA-molecules alone (The ssDNA-molecules are depicted as a sequence of red rods with the thiolated tail (C6)). From Figure 3.7 we notice that the three profiles exhibit similar time variations as the ones shown in Figure 3.6. More interestingly however, are the absolute variations among the different SAM-layers, for which, as a function of the presence of MCH, Cd decreases. The device functionalized with MCH 36 3 Two-Electrode Configuration alone possesses the smallest value of Cd whereas the device with no MCH exhibits the biggest one. This behavior is in agreement with the theory. According to the model of the differential capacitance explained in the introduction in fact, the functionalization layer can be modeled as a simple capacitance, Cmol , in series with the one coming from the diffused ion in solution, Cions . Making use of this model we can thus describe Cmol as: Cmol = 0 A/d (3.5) where A is the area of the electrode in contact with the solution, d the height of the molecular layer, and 0 the dielectric constant of vacuum and molecular layer, respectively. According to this equation and since A remains constant for all devices, Cmol can vary either because of changes in the height of the molecular layer or because of variations in the dielectric constant, . In the case studied in Figure 3.7, the change in the dielectric constant dominates. According to the literature in fact, DNA-SAMs are by far less packed than thiols-SAM [54] and thus, referred to our case, the ssDNA-SAM is much less ordered than the MCH-SAM. Because of the lack of order and the free volume in the DNA-SAM, water molecules can penetrate and interact with the DNA-molecules increasing the average dielectric constant of the molecular layer. In Figure 3.7, on the right, we tried to give a pictorial explanation of the suggested thesis. In case a), a SAM formed by MCH-molecules alone, there is absolutely no space for water molecules within the functionalization layer whereas in case c), a SAM formed by DNA-molecules alone, water can easily enter it and contribute to the total capacitance. We believe that the increase of the average dielectric constant, due to the water molecules, is the main cause of the increase in the value of Cd that we observe in Figure 3.7. We note at this point, that the height of the MCH-SAM with respect to the one of the ssDNA-SAM (≈ 1 vs 2.5 nm) should induce a decrease of Cd . The change however, happens to be more important and hence hides the information about the height of the molecular layer. At this point of our study we demonstrated that, following the value of the differential capacitance in time, we were able to discriminate among different functionalizations of the electrodes. However we continued our characterization of the two-electrode devices exploring another possible source of instabilities: attachment of multivalent ions on the electrode surface. It is known in fact, that multivalent ions can stick on gold surfaces altering in this way the Stern’s capacitance of the electrodes (see Appendix A). Passivation of the electrodes with a molecular layer is a diffused strategy in order to avoid this problem [65] and we wanted to test it on our devices. Possible irregularities in the SAM-formation in fact, could lead to missleading results in the following steps of the device development, i.e., immobilization of protein binders and detection of protein binding events. Thus we checked the two-electrode devices against different ionic buffers. In order to perform this test we used a passivation-layer formed by a SAM of C14-thiols. This molecular layer is known to pack very densely and in a very ordered manner on gold, passivating very well the surface [54]. In Figure 3.8 we compare the behavior of Cd , measured in KCl 10 mM, as a function of a 4 h exposure to three different buffers used in our group for protein analysis: PBS, TE and KH2 PO4 . We proceeded as follows: first we functionalized the electrodes with a C14-SAM (immersion in Ethanol containing C14 1 mM overnight), second we measured Cd over time in KCl 10 mM for 5 h, third we fluxed in the channel one of the three buffer solution for 4 h and finally we repeated the measurement of Cd in KCl 10 mM. We repeated steps 2 to 4 for each buffer and we plotted the results in Figure 3.8. 3.3 Time Behavior of the Two-Electrode Setup 37 Fig. 3.8. Absolute capacitance change as a function of time measured in KCl 10 mM for the same electrodes functionalized with a C14-SAM after the flux in the channel of different buffer-solutions for 4 h. Black: freshly prepared C14-SAM; blue: after flux of KH2 PO4 ; green: after flux of PBS ; red: after flux of T E at pH = 9. From the curves in Figure 3.8 we can see, that the flux of the different buffers induces variations in Cd . This counterintuitive behavior can be explained if we assume that the SAM-layer of the C14-molecules has some irregularities and thus that the ions present in the different buffers can stick on the gold surface. If this happens, the variations arise from the variations in the Stern’s capacitance (see details in Appendix A). Ions can in fact stick on the gold electrodes and thus alter the first layer of the double layer capacitance, C DL , which is in parallel with Cmol , the sensing element upon which our device is based. In Figure 3.9 we try to give a pictorial view of the phenomenon we believe the cause of the variations present in Figure 3.8. In this cartoon we emphasize the irregularities of the SAM (empty gold areas) and thus we clarify the contribution of the multivalent ions (purple and grey circles), that stick on the gold surface. After this first experiment we tried to improve the quality of the SAM cleaning the electrodes surface before functionalization. We tested several techniques: Reactive Ion Etching (RIE), chemical cleaning and electrochemical stripping but we always observed changes of Cd upon buffer variation. At this point we considered to perform differential measurements. Theoretically in fact, measuring the variation of Cd with respect to an identical electrode, permits to measure all kind of recognition events without any concern about time stability and ions adsorption. Unfortunately however, we were not able to produce electrodes with identical behaviors, both concerning the time stability and the absolute value of the differential capacitance. An intrinsic variability of the fabrication process lead to unpredictable behaviors of the microelectrodes. This problem is well known in literature [35] and is the main reason why differential measurements are not performed. Nevertheless we still tried to carry out some measurements 38 3 Two-Electrode Configuration Fig. 3.9. Cartoon that emphasizes the irregularities of the SAM (empty areas within the green bars) and clarifies the contribution to Cd given by the multivalent ions (purple and grey circles), that stick on the gold surface after the flux of different buffers in the channel. with the two-electrode configuration and check its ultimate capabilities. For this reason we tried to measure protein-detection immobilizing the protein binder of interest via DDI. In order to avoid the problems of the ions adsorption and, as a consequence, the related variations of the capacitance, we decided to use always the same buffer throughout the experiment. In this way we hoped to measure variations of Cd induced only by the variations in the molecular layer on the electrode and not by other sources. The buffer of choice was T E NaCl 1 M. It is a good buffer for DNA-hybridization because its high ionic strength favors the DNA-hybridization and thus the DDI process. The results are summarized in the next section. 3.4 Protein Detection 39 3.4 Protein Detection As mentioned in Section 3.3, we tested the capabilities of the two-electrode configuration applying our device to protein-detection. In order to perform this series of experiments we used streptavidin-DNA conjugates that are covalently linked together. These molecules were produced in the laboratories of our collaborator, Dr. Fruk at the Karlsruhe Institut of Technology, Germany. The ssDNA has the complementary sequence of the ssDNA attached to the gold electrode and thus bind to it via DNA base pairing. This process is at the base of DNA directed immobilization (DDI), the immobilization technique used in our laboratory in order to create protein patches starting from the simpler immobilization of ssDNA [8]. In Figure 3.10 we show a pictorial view of DDI. Fig. 3.10. Cartoon of the DDI-process for a SAM of ssDNA (shown on the left) after incubation with the ssDNAstreptavidin conjugates (shown on the right). The ssDNA of the conjugate has the complementary sequence of the immobilized ssDNA on the surface. The conjugates were produced in the laboratories of our collaborator, Dr. Fruk at the Karlsruhe Institut of Technology, Germany. We performed the experiment as follows: first we functionalized the electrodes with a LD ssDNASAM + MCH (see Figure 3.10a) and measured Cd in KCl 10 mM for 5 h. Second we fluxed in the channels a solution of T E NaCl 1M containing a concentration of 500 nM of the the Streptavidin-DNA conjugates for 1 h (see Figure 3.10b). Finally we measured again Cd in KCl 10 mM for 5 h. The results are shown in Figure 3.11. From Figure 3.11 we notice that the two profiles, blue and red, are very different even though the buffer we used, T E NaCl1 M, remained unchanged. After the reaction, i.e., hybridization of the DNA-molecules, we observe an increase in capacitance, which is in agreement with the existing literature. This increase is a bit counterintuitive since the final SAM is composed by dsDNA conjugated with Streptavidin and hence is higher than the original ssDNA-SAM. The increase in height, which amounts to roughly 4 nm (2 nm for the hybridization of the DNA and 2 nm for the streptavidin), should in fact induce a decrease in Cd , which, however, we do not measure. A possible explanation, suggested by Carrara et al. [60], is that the protein layer which forms at the top of the SAM changes completely the charge distribution of the ions around the biomolecules and induces an increase of the measured capacitance. In order to check whether we could further stress our system, we tried to bind a third molecule, which, we thought, should have decreased the 40 3 Two-Electrode Configuration Fig. 3.11. Cd vs t profiles as a function of DDI and protein-protein binding in a solution of KCl 10 mM. Blue: electrodes functionalized with a SAM of ssDNA + MCH; red: Cd after hybridization of the DNA-molecules with the complementary ssDNA conjugated with streptavidin-molecules; green: Cd after the interaction of the streptavidin with a solution containing biotinilated aGFAP-molecules, the antibodies for GFAP. total capacitance. Towards this goal we fluxed in the channel a solution of T E NaCl 1M containing, this time, biotinilated antibodies for Glial Fibrillary Acidic Protein (GFAP) in a concentration of 2.5 µg/mL. The antibodies for GFAP (aGFAP) are loaded onto the DNA-Streptavidin SAM through the high affinity (≈ 10−15 M) between Streptavidin and biotin. Finally we measured again Cd in KCl10mM. GFAP has been recognized as a biomarker for gliomas cancer, is expressed in the central nervous system and represents a target for the early diagnostics and the therapeutic monitoring of patients. In our ideas the antibodies for GFAP should decrease Cd increasing the total height of the SAM without profoundly changing the charge distribution of the biological layer. The green points in Figure 3.11 represent the experimental results. As expected, the addition of a second protein has the effect to decrease the capacitance of the system. From the time behavior of the Cd signals, we can try to extrapolate a general protocol in order to perform protein detection with the two-electrode detectors. First of all one has to measure for long time, at least 5/6 h. Secondly one must use the same buffer for all the functionalization steps in order to avoid missleading results. Third, with these pieces of information, one can make averages over many hours as a function of the electrodes functionalization. Following these three rules we built the graph shown in Figure 3.12 where we plot Cd as a function of the electrodes functionalization. The capacitance was calculated averaging the experimental results shown in Figure 3.11 over the last four hours of the experiments. In Figure 3.13 we further show similar results obtained by Carrara et al. in 2009 [60]. In their work the authors developed a device for protein detection based on the measurements of the differential capacitance. 3.4 Protein Detection 41 The device is composed by a couple of interdigitated electrodes and was tested on the recognition of Hepatocarcinoma marker SCCA from patient’s serum. Fig. 3.12. Cd measured in KCl 10 mM as a function of the electrodes functionalization. The values of the differential capacitances were computed averaging the experimental results shown in Figure 3.11 over the last four hours of the experiment. The error is the root mean squared deviation in the same time interval. In Figure 3.13 the capacitance values obtained by Carrara et al. immediately after the antibodies film formation, after BSA exposure and after SCCA exposure respectively are shown. BSA does not induce any change in the measured capacitance and demonstrates the specificity of the assay. From Figure 3.12 and 3.13 we observe that in both cases the capacitance increases after protein-binding. We know that this variation is proportional to the charge and the dimension of the protein under test and thus it is difficult to make a comparison of the absolute changes of Cd in the two different experiments. Nonetheless in both studies it sums up to roughly 10%. More precisely, in our experiment it is circa 8% whereas in [60] it reaches 16%. We note here that the capacitance measured by Carrara et al. is much bigger than the one measured with our setup. The reason is that, in their configuration, the authors use a couple of interdigitated electrodes with a big area whereas the surface of our electrodes is only 100x100 µm2 . The difference in the total area causes the difference in the absolute value of the capacitance. 42 3 Two-Electrode Configuration Fig. 3.13. Histograms from [60] where the authors compare the capacitance measured at the interface electrode/electrolyte as a function of the molecular layer on the surface of the electrode. First bar:film of antibodies; second bar:after incubation with albumine; third bar: after incubation with SCCA. 3.5 Problems of the Two-Electrode Setup and Possible Solutions 43 3.5 Problems of the Two-Electrode Setup and Possible Solutions In this final section we want to conclude the chapter reassuming the advantages and disadvantages of the two-electrode configuration examined so far. First of all we list the plus. The setup with two electrodes is easily integrable in more complex circuitry and the absence of a reference electrode greatly simplifies the geometry and the fabrication of miniaturized devices. Further, with the two-electrode configuration we were able to detect DNA-hybridization and, with several attentions in the experimental procedures, protein-protein interactions. Nevertheless we faced many problems. First, we noticed a strong dependence of the Cd signal on time and the passivation of the electrodes with a very ordered and thightly packed C14-SAM did not completely solved the problem. Second, we were obliged to change the experimental procedure and use the same buffer in all the functionalization steps in order to avoid misleading results induced by the absorption at the electrodes of multivalent ions. Third, the capacitance signal was overall unstable. We have not yet discussed this point but it is of great importance because we encountered it many times during our experimental work. In Figure 3.14 we show an example of such instabilities. Fig. 3.14. Cd measured in KCl 10 mM, over a period of 30 h and for a two-electrode-device functionalized with dsDNA-SAM. Time instabilities are clearly visible throughout the measured time-range. The graph reports Cd measured for electrodes functionalized by a dsDNA-SAM in KCl 10 mM over a period of 30 h. At the beginning, as observed in other experiments, Cd decreases but, after roughly 5 h, we observe a sharp increase in the capacitance signal which stops after 13 h. At this point a second decaying phase begins. This kind of behavior was observed for several detectors and is completely inexplicable unless we hypothesize that the voltage of the metal electrodes with respect to the solution can abruptly change and thus induce variations of Cd . After a long discussion about the origin of such instabilities, we 44 3 Two-Electrode Configuration concluded that the main problem of our two-electrode setup could be the absence of a proper reference electrode. This electrode is fundamental when performing electrochemical measurements in liquid and guarantees that the potential we applied across reference and working electrode does not undergo variation in time. Reference electrodes have the ability to equilibrate fast within an electrolytic solutions and, unlike metal electrodes, they behave very similarly to an ideal non-polarizable electrode, i.e., they do not undergo unpredictable potential drift caused by accumulation of charges at the electrode. For this reason we decided to upgrade our experimental setup introducing a third reference electrode. In the next Chapter we will present the results we obtained with the three-electrode configuration and the progresses we made beyond the two-electrode setup. Because of the dimension of the reference electrode, we were obliged to renounce to the microfluidic channel and perform the experiments in a circular pool with a diameter of 6 mm, which can host 100 µL of liquid. Although the final goal of our project has always been to develop a miniaturized system, abandoning the microfluidic channel was, at the time, necessary. The priority in fact, was to test whether the introduction of a third reference electrode could indeed stabilize the measurements and simplify data collection. If this was the case, we would have tackled the problem of the microfluidic channel in a successive step. 4 Three-Electrode Configuration In this Chapter we first characterize the three-electrode device as we did for the two-electrode one. In the second part we measure DNA-hybridization performing experiments as a function of the applied potential and we test the stability vs time. Eventually we show some measurements of DNA-hybridization in real time and apply Langmuir kinetics to describe it. 4.1 Electrical Characterization In Figure 4.1 we show the connections among the different electrodes in the three-electrode setup. The WE and CE are microfabricated gold electrodes (see chapter 2) while the RE is a classical Ag/AgCl pellet electrode. Fig. 4.1. Scheme of the connections of the three-electrode setup. The potential is applied across WE and RE whereas the current is measured across WE and CE. As in a normal electrochemical setup, the potential is applied across WE and RE whereas the current is measured across WE and CE. The only difference with standard electrochemical setups is that the RE, in our configuration, is directly immersed in the pool without any glass-frit. This element is often present in 46 4 Three-Electrode Configuration standard electrochemical setup and its role is to separate the experimental environment from the solution in contact with the RE and, at the same time, permit the exchange of ions. The absence of the glass-frit in our configuration would raise problems in case we were willing to compare our measurements with results obtained using normal electrochemical setups. Therefore we performed a calibration of our RE with respect to a standard reference electrode (sRE): Ag/AgCl in a solution of saturated KCl. Practically, we measured the open circuit potential (OCP) of our RE with respect to the sRE in different saline solution of KCl and NaCl. The open circuit potential is the potential across WE and RE, for which no current flows across WE and CE. We measured the OCP in 4 different solutions of KCl and NaCl for 1 minute and we then computed averages and root mean squared deviations. In Figure 4.2 we show one example of the measurements performed in the case of KCl. Using the values of the OCP in the different saline solutions (see Table 4.1), one can relate the potentials measured in our setup to the standard Ag/AgCl-RE using the following relationship: ∆VWE−sRE = ∆VWE−RE + ∆VRE−sRE (4.1) where ∆VWE−sRE is the potential difference which would exist between our WE and the sRE while ∆VWE−RE and ∆VRE−sRE are the tensions measured across WE vs RE and RE vs sRE, respectively. Fig. 4.2. Values of the OCP across our Ag/AgCl-RE and a standard RE, Ag/AgCl in saturated KCl solution, for 4 different concentrations of KCl. In the graph we use the nomenclature of Equation 4.1. Red: KCl 100 µM; blue: KCl 1 mM; green: KCl 10 mM; black: KCl 100 mM. The OCP-value was measured for 1 min. Average-values and root mean squared deviations were computed in this time interval and are listed in Table 4.1. 4.1 Electrical Characterization salt concentration ∆VRE−sRE (V) 47 salt concentration ∆VRE−sRE (V) KCl 100 µM (0.166 ± 0.004) NaCl 100 µM (0.152 ± 0.002) KCl 1 mM (0.166 ± 0.001) NaCl 1 mM (0.155 ± 0.001) KCl 10 mM (0.136 ± 0.002) NaCl 10 mM (0.132 ± 0.001) KCl 100 mM (0.087 ± 0.001) KCl 100 mM (0.088 ± 0.001) Table 4.1. Average-values and root mean squared deviations of the values of ∆VRE−sRE computed from the OCP measurements in different salt solutions. The averaging was performed over a time interval of 1 min. As already explained, the data reported in Table 4.1 are important in the case we should compare the potential values obtained in our setup with the values of another experiment, performed with a standard RE. Since we will not have this need in the rest of the thesis, we will not use them. Nonetheless we decided to include the calibration of our RE with respect to the sRE because it could be useful for the readers. After calibration, in analogy with the two-electrode setup, we performed the characterization of our device vs frequency. We thus acquired current measurements in KCl 10 mM (concentration selected from previous measurements) at different frequencies and plotted the relative results (see Figure 4.3). The applied voltage had a root mean squared amplitude of 10 mV. Fig. 4.3. Root mean squared current, Irms , plotted as a function of the applied frequency, f , for a solution of KCl10mM and an applied potential, Vrms = 10 mV. From the graph we notice that, as happened for the two-electrode configuration, the current measured in the three-electrode devices is dominated by the double layer capacitance at low frequencies (below 1 kHz) whereas, above this threshold, the system starts to be dominated by the ionic resistance of the 48 4 Three-Electrode Configuration solution. In analogy with the two-electrode devices we assume that, at higher frequencies, the signal follow the capacitive behavior given by the stray capacitance. Unfortunately, with the bipotentiostat we cannot measure at frequencies higher than 2 − 3kHz and thus we could not measure it directly. After this calibration we started to challenge our device with the detection of DNA-hybridization. The value of Cd was derived from the linear fit (see Equation 3.3) of the Irms -values in the range (100−400)Hz as explained in the previous Chapter 3. These measurements will be described in the next sections. 4.2 DNA Hybridization as a Function of the Applied Bias Potential 49 4.2 DNA Hybridization as a Function of the Applied Bias Potential According to the Stern model for the characterization of the double-layer capacitance, C DL , explained in Appendix A, a minimum of Cd exists for a certain applied potential. At this potential the surface charge density of the electrode counterbalances exactly the charge of the ions attached to the surface and thus the contribution to Cd given by the Stern layer becomes negligible (see Appendix A for more details). In our system we passivate the surface of the electrode with a densely packed SAM and thus we should not observe this minimum. Nevertheless in the previous Chapter we argued that changes of Cd upon immersion of our device in different buffers, could be due to absorption of multivalent ions on the surface of the electrodes. In order to test this idea we performed several experiments of DNA-hybridization as a function of the DC-potential applied at the WE with respect to the RE. In this way we had a double opportunity. On one hand we could test whether the two-electrode devices are influenced by the Stern’s layer and, at the same time, we had the chance to measure the potential at which the minimum of Cd appears. This value would represent another possible quantity of interest and its changes upon DNAhybridization could give us supplementary information to be used in DNA-detection. We proceeded as follows: first we functionalized the WE with a ssDNA SAM (see Section 2.5) and we filled the pool with the measuring solution, KCl 10 mM. At this point we measured the open circuit potential (OCP), the potential between WE and RE, at which no current flows across WE and CE. As a function of the OCP we then chose the potential range for our measurement. In general we measured a maximum potential range of 1V around the OCP-value. After several trials in fact, we noticed that too high potentials can lead to electrochemical desorption of the thiols. Because of this problem we tried to limit the scan of the potential in order to avoid desorption. After this first measurement as a function of the DC applied potential, we changed the solution in the pool adding T E NaCl 1 M containing the complementary ssDNA at a concentration of 1µM and we stored the device in dark for 1.5 h. In the end we repeated the initial measurement in KCl 10 mM using the same potential range. One set of experimental data is shown in Figure 4.4. From Figure 4.4 we notice, that, both for ssDNA and dsDNA, Cd does not present the sharp minimum foreseen by the Stern’s theory. On one hand this tells us that the feared absorption of multivalent ions on the electrodes is not very problematic and, by the other hand, that we cannot rely on it in order to perform detection of DNA-hybridization. Nevertheless we see from the figure that the capacitance decreases when we hybridize the SAM on the electrode and this agrees with the previous experiments performed with the two-electrode setup and with literature [57, 58, 35, 50]. From Figure 4.4 we can further compute the decrease of Cd from ssDNA to dsDNA as roughly 10% throughout the explored potential range. On the very right margin of the plot, for a value of VBias = 0.2 mV, we notice a sharp increase of the capacitance. This sharp change of Cd can be related to thiols desorption and thus we stopped the measurement at this applied potential in order to avoid SAM damaging. At this point we wanted to be absolutely sure that the decrease of Cd was due to DNA-SAM hybridization. As direct control of DNA-hybridization we thus performed AFM measurements. The three-electrode configuration in fact, gives us the opportunity to use AFM and test directly the height change of the SAM upon DNA hybridization. This possibility is granted by the fabrication of the devices. In the threeelectrode detectors we added an isolation-layer that limits the area of the electrodes in contact with the 50 4 Three-Electrode Configuration Fig. 4.4. Cd vs VBias as a function of DNA-hybridization in a solution of KCl 10 mM. red: electrodes functionalized with a SAM of ssDNA + MCH; blue: Cd after hybridization of the DNA-molecules with the complementary ssDNA. solution (see 2.2). Due to this delimitation we know exactly where the electrode is functionalized and thus we can choose an area of the electrode where to perform the AFM-measurement. In Figure 4.5 we marked the area in which we decided to perform the AFM measurements. Fig. 4.5. contours of the area of the working electrode within which we chose to perform the shaving experiment with AFM. In particular, with the AFM-tip operating at high pressure, we shaved away SAM-molecules from a defined area (in our case 1 µm2 ) in order to measure the height profile of the SAM with respect to the 4.2 DNA Hybridization as a Function of the Applied Bias Potential 51 underneath gold surface [66]. We scanned at a high force (≈ 100 nN) with the AFM-tip in contact with the surface. The scanning at high load induces stripping of the molecules and the scanned area results completely shaved. After the first scan one can return to low force (to avoid SAM-compression) and measure a bigger region of the sample that includes the shaved one (see for instance the inset of Figure 4.6). From the height of the hole information about the conformation of the DNA-molecules present in the biomolecular-layer can be extracted. The images we collected for ssDNA and dsDNA together with the histograms of the height distributions are shown in Figure 4.6 and 4.7, respectively. As already explained, these measurements were performed after shaving, in contact mode and at low force in order to avoid compression of the SAM. From the images we notice that the measurements were not very clean. This is principally due to two reasons: first we performed the measurements on the same samples on which we did the EIS-experiments and secondly because, in order to keep constant the solution, we did the shaving and imaging in KCl 10 mM. This buffer is not the ideal one and it is probable that some residues of the shaving remains at the border of the hole and form the bright areas we notice in the images. Nevertheless these images are very informative and can give us the proof of whether the DNA-hybridization took place. Fig. 4.6. Histogram of the heights of the image shown in the inset. The shaving experiment was performed for electrodes functionalized with ssDNA + MCH. Blue line: fit of the histogram performed with gaussian functions. From the two histograms shown in Figure 4.6 and 4.7, we can distinguish two distinct peaks. The first one, on the right, is the height of the SAM-carpet whereas the second one, on the left, corresponds to the height of the hole. In Figure 4.6, i.e., for the ssDNA SAM, the two peaks are a bit superimposed whereas in Figure 4.7, i.e., for the dsDNA SAM, the two peaks are well separated. From the distance between the two maxima we can compute the height of the molecules, that build up the SAM, and thus understand 52 4 Three-Electrode Configuration Fig. 4.7. Histogram of the heights of the image shown in the inset. The shaving experiment was performed for electrodes functionalized with dsDNA + MCH. Blue line: fit of the histogram performed with gaussian functions. whether they are in agreement with the expected heights for ss- and dsDNA. In order to do it we fitted the histograms with gaussian functions. The parameters of the gaussian function can be extrapolated from the following equation: −(x − x0 )2 (4.2) 2d2 where A is the amplitude of the function at x = x0 , x0 the center of symmetry of the function while d f (x) = Aexp carries information regarding the width of the bell. In the case of dsDNA we were able to fit the experimental point with the sum of only two gaussians whereas for the ssDNA we had to use a third gaussian. The reason is the bright area in the inset of Figure 4.6 located at the border of the hole. This dirtiness is represented in the histogram and thus we need another Gaussian in order to fit it. In Figure 4.6 and 4.7 the two fits are drawn in blue while the fitting parameters are summarized in Table 4.2. From the position of the maxima obtained from the two fits we compute the following SAM-heights for ssDNA and dsDNA, respectively: difference ssDNA dsDNA |x1 − x2 | (2.38 ± 0.07) nm (4.87 ± 0.02) nm The difference we measure between ssDNA and dsDNA amounts to (2.49 ± 0.09) nm and is in agreement with the expected height difference upon hybridization of a 22-bases ssDNA at low density and 4.2 DNA Hybridization as a Function of the Applied Bias Potential Fitting parameter ssDNA 53 dsDNA A (400 ± 31) counts (2496 ± 10) counts B (516 ± 30) counts (598 ± 10) counts / C (1119 ± 23) counts x1 (−3.85 ± 0.06) nm (−5.02 ± 0.01) nm x2 (−1.47 ± 0.01) nm (−0.15 ± 0.01) nm x3 (−0.98 ± 0.06) nm / d1 (0.90 ± 0.04) nm (0.74 ± 0.02) nm d2 (0.48 ± 0.02) nm (0.73 ± 0.01) nm d3 (1.56 ± 0.03) nm / Table 4.2. Fitting parameters of the gaussian equation for the shaving experiments of ssDNA- and dsDNA-SAM. In the case of dsDNA we were able to fit the experimental points with the sum of only two gaussians whereas for the ssDNA we had to use a third gaussian. The reason is the bright area in Figure 4.6 located at the border of the hole. This dirtiness is represented in the histogram and requires another gaussian for the fitting. The meaning of the parameters can be extrapolated by Equation 4.2 in a solution of KCl 10 mM [52]. Thus, after the AFM-measurements, we were absolutely sure that the difference of Cd , that we measured in Figure 4.4, came from the DNA-hydridization of the SAM. We note here that the presented shaving measurements were obtained on the very same sample on which we performed the EIS-experiment. This note is of remarkable importance since, with the threeelectrode setup, we can directly check the different steps of the DNA detection immediately after the electrochemical measurements. In literature there are already examples of AFM measurements linked with EIS-experiment but they are always performed on dedicated samples [44]. Further, the samples used for the AFM study are normally very particular and employ the so called template stripped gold. These samples are obtained via stripping from a very flat surface, like Mica or Silicon-wafer, and thus present very flat and clean gold surfaces. In our experiment, on the contrary, the AFM measurements were performed on the same sample which we investigated with EIS and thus, can directly confirm or deny the electrical measurement. We think that this aspect could be of particular importance in the future developing-phase of the device when we will apply it on more complex tasks, as protein detection on a protein monolayer. 54 4 Three-Electrode Configuration 4.3 Time Behavior of the Three-Electrode Setup Knowing the time instabilities shown by the two-electrode devices (see Section 3.3), we tested the threeelectrode setup vs time. In analogy with the two-electrode configuration we measured Cd as a function of different electrode functionalizations over a time interval of 5 h. In Figure 4.8 we plot the obtained results for three passivation-layers: MCH, ssDNA and dsDNA. For the sake of clarity and in order to avoid superimposition of the experimental points, the three plots are artificially shifted by 5 min one from the other. As already done for the two-electrode setup, we plot the percentage change of Cd instead of the absolute values because, in this way, the comparison results easier. Fig. 4.8. Normalized Cd -change vs time as a function of three different functionalization-layers, in a solution of KCl 10 mM. red: electrodes functionalized with a SAM of MCH; blue: electrodes functionalized with ssDNA + MCH; green: electrodes functionalized with dsDNA + MCH From Figure 4.8 we notice immediately that the time decay characterizing the two-electrode configuration disappears completely. The value of Cd , for the three tested passivation-layers, varies of a maximum of 2 % throughout the experiment and, most important, does not decay with time. In order to highlight the striking difference between the three-electrode device with respect to the two-electrode one, we plot in Figure 4.9 the same figure shown in the Section 3.3 which we used to describe the time behavior of the two-electrode detectors. In Figure 4.9 we thus show Figure 3.6 with the addition of the green plot of Figure 4.8, i.e., WE passivated with dsDNA, extended to a time interval of 9 h. From Figure 4.9 we can appreciate the stability of the three-electrode detectors in respect to the twoelectrode ones. The Cd -signal does not undergo decay over time and is stable from the very beginning of the measurement. The latter feature will be of considerable importance in the last part of this chapter, 4.3 Time Behavior of the Three-Electrode Setup 55 Fig. 4.9. Figure 3.6 with the superimposition of the green plot in Figure 4.8. Red: two-electrode-setup, bare electrodes; blue: two-electrode-setup, SAM of C14 thiols, green: two-electrode-setup, SAM of ssDNA + MCH; black: threeelectrode-setup, SAM of dsDNA + MCH. when we will study the kinetics of the DNA-hybridization. We note here, that the measured time stability was even longer than 9 h. We tested different samples overnight and the variations of Cd were always less than 3 % of the initial value and, we remark it again, no decay trend could be observed. In Figure 4.10 we show one of this measurement as an example. In this Figure we plot the absolute value of Cd measured at a WE functionalized with dsDNA over a time interval of 11 h. The average value of Cd and its standard deviation over this time interval are: Cd = (0.503 ± 0.004) nF. At this point we started to test whether, with the three-electrode configuration, we were able to detect DNA-hybridization. We proceeded as explained in Section 2.5. First we functionalized the WE with a mixed layer of ssDNA and MCH and then measured Cd vs time (2 h). After DNA-hybridization we repeated the measurement of Cd and we compared the two results. In Figure 4.11 we show a representative example among the several experiments that we performed. As we can see from the graph, the expected decrease of Cd upon DNA-hybridization cannot be measured within the experimental errors. This problem puzzled us several weeks until we realized that, probably, the SAM did not form any more on the gold electrodes. In order to test it we performed again AFM-shaving and indeed we confirmed the absence of the biological layer. Along with the test done with the AFM we tested the absence of the SAM also with EIS. We did it measuring Cd before and after the procedure for SAM-formation. In Figure 4.12 we show the comparison. From Figure 4.12 it becomes clear that, at this stage of the project, we were not able to form a densely packed SAM on the surface of the electrode. The reasons can be many but we believe that there are two 56 4 Three-Electrode Configuration Fig. 4.10. Cd measured in KCl10mM using a three-electrode setup with a WE functionalized with dsDNA over a time interval of 11h. The average value of Cd and its standard deviation over this time interval are: Cd = (0.503±0.004)nF. Fig. 4.11. Cd measured in KCl 10 mM vs time as a function of DNA-hybridization. Red circles: WE functionalized with ssDNA + MCH; blue circles: Cd after DNA-hybridization. 4.3 Time Behavior of the Three-Electrode Setup 57 Fig. 4.12. Cd measured in KCl 10 mM vs time as a function of SAM formation. Red circles: clean WE; blue circles: WE after the procedure for ssDNA-functionalization. main causes. The first one is the degradation of the gold we used for evaporation whereas the second one can be related to the optical resist we used in the isolation of the electrodes. The resist in fact, must be spinned on the whole samples and then selectively removed from the electrodes with an aligned lithography. If a thin film of resist remains on the electrode it can hinder thiols adsorption and thus the formation of the SAM. To continue our tests we decided to clean the surface of the electrodes with a plasma etching prior to DNA exposure. The parameters that we used in order to etch the first 2 nm of gold and expose in this way the clean metal, are listed in Table 4.3. Element Oxygen O2 Argon Ar Power (W) Flux (sccm) Pressure (mbar) DC Potential (V) Time (min) 20 35 30 1 · 10−1 150 1 20 −1 240 5 1 · 10 Table 4.3. Parameters of the plasma that we used in order to etch the first 2 nm of gold and expose in this way the clean metal. The plasma treatment was performed in the chamber of the machine for reactive ione etching (RIE). After the introduction of the etching procedure prior to the functionalization of the gold electrodes, we repeated the experiment of the DNA-hybridization. The results we obtained are shown in Figure 4.13. In Figure 4.13 the decrease of Cd upon DNA-hybridization is again evident and amounts to roughly 40 % of the starting value,i.e., the capacitance corresponding to the ssDNA. 58 4 Three-Electrode Configuration Fig. 4.13. Cd measured in KCl 10 mM vs time as a function of DNA-hybridization for a device treated with plasma etching in order to expose the cleanest gold surface possible. Red circles: WE functionalized with ssDNA + MCH; blue circles: Cd after DNA-hybridization. At this point of the project we had demonstrated to be able to measure DNA-hybridization through highly stable Cd measurements vs time, in a solution of KCl 10 mM. However, as we said before, this corresponds to an ionic strength too small to deal with real biological entities as DNA and, more importantly, proteins. We therefore decided to test our device in a buffer solution with KCl 100 mM, a salt concentration closer to the biological environment (200 mM). Again we decided to challenge our device with the detection of DNA-hybridization. We followed the usual procedure for SAM-formation and DNA-hybridization and we collected the capacitance signal in KCl 100 mM. Prior to SAM-formation we performed the plasma etching procedure in order to start with the cleanest gold surface possible. In Figure 4.14 we show the experimental results. Unfortunately the first data collected, shown in Figure 4.14, showed no variation of Cd upon exposure of the DNA-SAM to the complementary strands. Again we tested the presence of the SAM with AFM but, because of the high roughness (≈ 5 nm) induced by the etching process, we could not draw a final conclusion. For this reason, after several trials, we decided to skip the MCH passivation-step. In this way we hoped to increase the contribution of the DNA to the changes of Cd upon hybridization. Doing it we were aware that this was at expences of the order of the ssDNA-film. In Figure 4.15 we show the results we obtained in this way. Surprisingly, skipping the MCH post-treatment, we obtained a variation of the capacitance upon DNAhybridization of roughly 15 %. We repeated the experiment several times starting from gold surfaces from different evaporation batches and we demonstrated that this result was reproducible. We believe that the 4.3 Time Behavior of the Three-Electrode Setup 59 Fig. 4.14. Cd measured in KCl 100 mM vs time as a function of DNA-hybridization for a device treated with plasma etching in order to expose the cleanest gold surface possible. Red circles: WE functionalized with ssDNA + MCH; blue circles: Cd after DNA-hybridization. Fig. 4.15. Cd measured in KCl 100 mM vs time as a function of DNA-hybridization and with the absence of the MCH-molecules in the SAM. Red circles: WE functionalized with ssDNA; blue circles: Cd after DNA-hybridization. 60 4 Three-Electrode Configuration most probable explanation can be the variation in the Debye-length of the buffer solution. The increase of the KCl concentration from 10 to 100 mM in fact, causes a decrease in the Debye length of the solution from 3 to 1 nm. This decrease, on its turn, causes a big decrease of the volume in solution influenced by the charged DNA. We tried to depict this situation in Figure 4.16 where we give a pictorial explanation of what could happen lowering the Debye length. For simplicity we draw here a cross section of the assembly composed by gold film, MCH carpet, DNA-molecules and solution. Moreover we depict two different situations of the system: before and after DNA-hybridization. In Figure 4.16 the DNA-molecules are represented as red rods while the MCH molecules are shown as zig-zag lines in contact with the surface. From literature [52, 59] we know that the persistence length of ss- and dsDNA amount to p ≈ 1 nm and p ≈ 50 nm, respectively. In Figure 4.16 we tried to reproduce a realistic situation in which the ssDNA is composed by 7 small rods (the length of a 22-mer DNA-molecule is about 7nm corresponding, for ssDNA, to 7 segments differently oriented with respect to each other) and the dsDNA by a single straight rod. The ions screens the charges of the molecules and form, around them, the so called electrical double layer (dashed lines for the two cases, i.e., κ−1 = 1 and 3 nm). Beyond this layer the ions in solution do not feel the charged molecules and do not contribute to the differential capacitance. In order to understand how the difference in the Debye length can conceal the contribution of the DNA-molecules to Cd , let us remember the model introduced in the first pages of the thesis (Figure 1.4). In that model Cd can be approximated by the series of two capacitances, Cmol and Cions . Since the latter is very big the former one determines the value of Cd . On its turn Cmol is an average of the molecules in the volume of the biological layer, in this case, MCH, DNA and water. Variations in this volume induce variation of the measured Cd . At this point we must consider the case that we are studying. The concentration of the DNA-molecules on the surface is 4−5·1012 molecules/cm2 which corresponds to 4−5 molecules in an area of 100nm2 . This density implies that the DNA-molecules are separated with each other by a distance of about 5 − 6 nm. When the Debye length is of the order of 1 nm, the DNA-molecules are completely screened by the ions in the double layer and do not interact. Cmol is made up by the MCH-molecules and by the isolated DNA-molecules plus a water layer with a thickness equals to the Debye length (see Figure 4.16a). Hybridization of the DNA-molecules influence scarcely the total volume that induces variations of Cd (see Figure 4.16b) and thus the latter remains constant. When the Debye length is of the order of 3 nm on the contrary, the DNAmolecules interact with each other and the volume in-between is strongly influenced by them (see Figure 4.16c). Cmol is composed by the MCH-molecules, the DNA-molecules and by the volume in-between. Hybridization changes the height of this volume and thus its contribution to Cd (see Figure 4.16d). This hypothesis is still under investigation and requires further experiments in which the density of the ssDNA-molecules in the SAM must be varied. For the moment we accept it as a possible explanation and we continue to present the results obtained with the three-electrode setup. 4.3 Time Behavior of the Three-Electrode Setup 61 Fig. 4.16. Cross section of the assembly composed by gold film, MCH carpet, DNA-molecules and solution. The DNA-molecules are represented as red rods, the MCH molecules are depicted as zig-zag lines on the electrode surface. The sulfur atom is shown as a blue circle while the OH-group as an orange one. Because of the variation in the persistence length of the DNA-molecules after hybridization (from 1 to 50 nm), the ssDNA is composed by 7 small rods whereas the dsDNA by a single straight rod. Also we draw ssDNA with a diameter of roughly half the length of the diameter for dsDNA, as is should be for buffer solutions that completely screen electrostatic charges. The limits of the electrical double layer is shown as a dashed lines. (a) ssDNA-SAM, κ−1 = 1 nm; (b) dsDNA-SAM, κ−1 = 1 nm; (c) ssDNA-SAM, κ−1 = 3 nm; (d) dsDNA-SAM, κ−1 = 3 nm. 62 4 Three-Electrode Configuration 4.4 Calibration Curve of the Three-Electrode Detectors Since the absence of the MCH-molecules facilitates the detection of DNA-hybridization, we decided to take advantage of this and stress our detector in the limit of detection. We performed therefore a calibration of our device. Using as a buffer solution KCl 100 mM and avoiding post-treatment with MCH, we performed the calibration exploring the dynamic range of detectable complementary DNA concentration over 4 orders of magnitudes, from 100 pM to 1 µM. For each concentration we left the solution in contact with the WE for 1 h and then we measured the percentage change of Cd relative to the value at t = 0, corresponding to the electrode functionalized with ssDNA only. The results we obtained are shown in Figure 4.17. Fig. 4.17. Calibration curve of the three-electrode device. The response is shown as percentage capacitance variation at the WE as a function of the complementary DNA concentration in solution (KCl 100 mM). The device saturates at a concentration of the target DNA of 100 nM whereas, at 100 pM, the capacitance change is already roughly 9%. From Figure 4.17 we notice that our device saturates at a solute concentration of about 100 nM with a maximum capacitance percentage change of 25%. The measured saturation value was the same for different devices explored (three). Moreover, at 100 pM the Cd change amounts to roughly 9%, well beyond the experimental error (5% at present) and therefore easily measurable. We think that once we will have improved our experimental setup (especially the gold quality) the detection limit of these devices for DNA-hybridization will be pushed towards the f M-range. Using the data shown in Figure 4.17 one can further compute the affinity constant, KA = ko f f /kon , of the DNA-hybridization. If we suppose in fact, that the DNA-pairing on the gold surface can be described by Langmuir adsorption isotherm, we obtain the following equation that connects the variation of 4.4 Calibration Curve of the Three-Electrode Detectors 63 the quantity sensible to DNA-hybridization (in our case Cd − change), the varying concentration of the complementary DNA in solution (ccDNA ) and the affinity constant (KA ) [67, 68]: KA 1 + ccDNA KA Reorganization of Equation 4.3 in the form of a linear function leads easily to Equation 4.4: Cd − change = Cd − change MAX ccDNA (4.3) 1 Cd − change MAX a (4.4) KA = where Cd − change MAX is the maximum change of Cd as a function of the cDNA-concentration. In Figure 4.18 we show the obtained plot of ccDNA /Cd − change versus ccDNA and the obtained linear fit in a Log-Log scale. Using equation 4.4 we thus obtained an affinity constant for DNA-hybridization KA = 5 · 109 M −1 . This value is in agreement with similar experimental results [67, 69]. Obviously the assumption we had to do in this part is that the DNA-hybridization follows a Langmuir kinetics. More on this topic can be found in the next section where we show direct measurements of DNA-hybridization kinetics. Fig. 4.18. Concentration of the complementary DNA in solution divided by the percentage change of Cd as a function of ccDNA . The graph is plotted in a Log Log scale and the linear fit of the experimental points is shown as a blue dashed curve. From the intercept of the linear function at ccDNA = 0 one can compute the association constant, KA , using Equation 4.4. 64 4 Three-Electrode Configuration 4.5 Kinetics of DNA Hybridization The time stability of the three-electrode setup opened up the possibility to study the kinetics of DNAhybridization in real time, which is of critical importance for biosensor design and optimization and for the efficient collection of genomic data [70]. Biologically relevant phenomena as hairpin folding, which plays a key role in biological functions as regulator of gene expression [71], or DNA single base pair flipping, fundamental in the interaction between DNA and DNA-repairing enzymes [71], can be in fact discriminated through kinetic and dynamic studies of DNA-hybridization. Many techniques have been employed in order to study DNA-hybridization kinetics. Among them we cite TIRF [70], FRET [59] and SPR [68]. EIS-experiments have also been carried out [67] but never in-situ. In their work, Li at al. reported that in-situ kinetics study experiments were hampered by the instability of their measured signals. They studied instead the variation of the charge transfer resistance ex-situ, hybridizing the DNA sample first and then transferring it in a solution of 5 mM Fe(CN)3−/4− and KCl 0.1 M. This procedure 6 slows down the experiments and limits the number of experimental points. Moreover the samples must be dried at each change of solution and this could lead to artifacts in the measurement. For all this reasons an in-situ approach would be preferable. Using our three-electrode detector we demonstrated to be able to follow DNA-hybridization in-situ. In order to do it we operated as follows: we measured Cd at the ssDNASAM functionalized WE, verified that the signal was stable, then changed the solution in the experimental pool adding a known concentration of complementary DNA. The buffer used for this experiment was a solution of KCl 100 mM in which we initially dissolved 1 nM complementary DNA. We used here the higher ionic strength (100 mM instead of 10 mM) to favor the hybridization process [51]. To follow hybridization kinetics, we initially measured at a rate of 4 measurement/min for 15 min, then we slowed down to 1 measurement/min for about one more hour. The results obtained are shown in Figure 4.19. In order to get insights into the DNA hybridization kinetics, we fit our data with the well-known Langmuir adsorption kinetics. In this theory adsorption can be described as a reversible process between adsorbent and adsorbate (or solute): A + B ← (kon ; ko f f ) → AB (4.5) where A is the solute, B is the adsorption site on the surface, AB is the complex and kon and ko f f are the adsorption and desorption rate constant, respectively. The relevant variable is the fraction of sites occupied at time t, that we denote by ϑt . The forward adsorption rate is first order with respect to the concentration of the solute at time t, ct , and with respect to the fraction of available adsorption sites, (1 − ϑt ). The desorption rate is first-order with respect to ϑt . The total adsorption rate can be expressed as: dϑt = kon ct (1 − ϑt ) − ko f f ϑt (4.6) dt Equation 4.6 is known as Langmuir kinetics. Following the procedure of Liu and Shen [72], the variation of ϑt over time can be also expressed as a function of ϑe , the value of ϑ at equilibrium, and two properly defined rate constants k1 and k2 , that in turn depend on Kon , Ko f f and the adsorbate dosage (see the paper for more details): 4.5 Kinetics of DNA Hybridization 65 Fig. 4.19. Study of the kinetics of DNA-hybridization via differential capacitance measurements. The WE of the threeelectrode setup was functionalized with a ssDNA SAM and its capacitance vs time was measured in KCl 100 mM. After 30 minutes we exchanged the solution in the pool with one having the same KCl concentration plus 1 nM of the complementary-strand DNA. The green and blue lines represent best-fit models based on first- and second-order Langmuir absorption kinetics, respectively. dϑt = k1 (ϑe − ϑt ) + k2 (ϑe − ϑt )2 (4.7) dt In this way, the adsorption rate equation is clearly written as the sum of a first order and a second order contribution. According to this new parametrization, it appears that the Langmuir kinetics is a hybrid rate equation with a variable reaction order of 1 − 2, and each term has a weight that depends on the relative magnitude of ϑe with respect to k1 /k2 . Langmuir kinetics has been applied by many groups to study DNA hybridization on solid surfaces. Peterson et al. [73] showed that a first order Langmuir model cannot satisfactorily describe their kinetics data. More recently, in a theoretical work, Wong and Melosh [74] studied DNA-hybridization as a function of DNA surface density and found that, at low DNA surface density (i.e. in the same conditions as ours), a first order Langmuir kinetics was sufficient to fit their data. However, they make the assumption that kinetics is reaction-limited, i.e., that target transport to the surface occurs much faster than target hybridization. This condition is not fully satisfied in our experiment since, if we compute the number of complementary DNA strands in a cube with a base area of about 100 nm2 (the average area for each DNA molecule on the surface of a low density DNA SAM) we find that the proportion between complementary DNA strand in solution and immobilized DNA on the surface is 1 : 106 at the working concentration of 1 nM complementary DNA. In other words we are in a situation where target transport cannot be ignored and probably, a second order rate equation is more appropriate to fit the experimental data. Given all these considerations we applied both models to our system. In Figure 66 4 Three-Electrode Configuration 4.19 we show both fits for comparison, the blue and green lines representing the fits using the second and the first order rate equation, respectively. The equations we used for fitting can be easily derived from the differential equations and under the assumption that the capacitance change is directly related to the number of occupied binding sites on the surface. The two equations (valid for t > t0 = 30 min) read: Cd = CdsDNA + C0 exp(−k1 t) (4.8) C0 (4.9) 1 + C0 k2 t is the final Cd after complete hybridization, C0 the difference between the capacitance Cd = CdsDNA + Where CdsDNA values of the initial ssDNA SAM (C ssDNA ) and the final dsDNA (CdsDNA ), k1 and k2 the first and second order rate constants, respectively. In order to perform the fitting in Figure 4.19 we further set (CdsDNA + C0 ) = C ssDNA where C ssDNA is another fit parameter for t < 30 min. As we can see from Figure 4.19, the second order rate kinetics fits better our results at the beginning and at the end of the hybridization process even though the data could be satisfactorily interpreted using the first order rate kinetics as well. Eventually we cannot exclude, for the conditions we used, to be in a situation where the transition from first order to second order Langmuir kinetics takes place. The parameters obtained from our fitting procedures are summarized in Table 4.4. Moreover, the overall DNA hybridization time derived from our measurements, = 1/k1 = (6.4 ± 0.2) min, is in good agreement with existing SPR data for hybridization in solution, at experimental conditions (i.e. ionic strength, DNA target concentration in solution) very similar to ours [14]. After this first proof of principle, we will continue the work on DNA-hybridization in real time. We know very well in fact, that this kind of data are very important for the future development of DNA microchips. In the last years many attempts in order to speed up the hybridization kinetics of DNA on solid surfaces have been performed [75]. The researchers tried to use microfluidic-networks in order to achieve this goal and a simple setup in order to measure DNA-hybridization could have a big impact in this field. We believe that an electrochemical setup would be best suited. Fitting parameter 1st order Langmuir kinetics 2nd order Langmuir kinetics C ssDNA (1.301 ± 0.002) F (1.301 ± 0.002) F C0 (0.229 ± 0.002) F (0.254 ± 0.003) F CdsDNA (1.072 ± 0.004) F (1.047 ± 0.005) F k1 (0.156 ± 0.006) min−1 / k2 / (0.89 ± 0.06) (F · min)−1 Table 4.4. Parameters of Equations 4.8 and 4.9 obtained through the fitting of the Cd -profile shown in Figure 4.19. 4.6 Passivation with Molecules of Thiolated Ethylene Glycol 67 4.6 Passivation with Molecules of Thiolated Ethylene Glycol We remind at this point of the description of the device that the calibration curve of our detector was obtained avoiding the use of MCH in the formation of the biological-layer on the surface of the electrode. We know however, that this absence would certainly lead to very big problems of unspecific absorption if the detector were used in a real biological sample. In a complex matrix like blood-plasma or urine in fact, unspecific absorption is the biggest hurdle to take in order to apply the detector successfully [76]. In literature a big concern has been given to this problem and many studies show that, in order to avoid unspecific absorption, a passivation-layer formed by bio-repellent molecules must be applied. In these works many authors suggest the use of oligoethyleneglycols as bio-repellent molecules [13, 8, 77] and thus we tried to use them as well. Practically we substituted the MCH-molecules with thiolated oligoethylene-glycols (TOEG6) and challenged our sensor upon detection of DNA-hybridization. With this experiment we wanted to test whether we were able to detect DNA-hybridization using TOEG6 as biorepellent molecules. In Figure 4.20 we show the collected results. Fig. 4.20. Cd measured in KCl 100 mM vs time as a function of DNA-hybridization and with the presence of the TOEG6-molecules in the SAM. Red circles: WE functionalized with ssDNA + TOEG6; blue circles: Cd after DNAhybridization. From Figure 4.20 we note that, using TOEG6 instead of MCH, we are able to detect DNA-hybridization and that the Cd -change passing from ssDNA to dsDNA amounts to roughly 6%. This decrease of the differential capacitance is smaller than the one measured without the MCH passivation layer but still detectable. We note at this point that, in order to reduce the absolute magnitude of the errors shown in Figure 4.20, we applied block analysis. Block analysis permits to select the correct number of statistically 68 4 Three-Electrode Configuration independent measurements which, in our case, amounts to 4 independent sets (4 sets, each containing 50 periods of the collected AC current-signal (see chapter 2 for the details of the performed measurement)). In this way we could reduce the errors by a factor 2. At this point of our research we were ready to apply our device for the detection of protein molecules in solution using the technique of DNA directed immobilization (DDI). Unfortunately the problems with the gold evaporation worsened and we were not able to perform these measurements. As soon as the gold quality will improve we will proceed along this direction. 4.7 Major Achievements of the Ph.D. Work In this final section we would like to summarize the major findings obtained during this Ph.D. project with respect to the state of the art in the field of bio-detectors based on electrochemical impedance spectroscopy. First, we demonstrated that the stability of the three-electrode configuration with respect to the one of the two-electrode setup is definitely better. This fact is not a new concept. From text books in fact, we know that the presence of a third electrode enhances the stability of the electrochemical measurements giving the possibility to reduce the current that flows through the RE [65]. Nevertheless a direct comparison of the two configurations in the context of EIS experiments was not present in the literature. The choice of a two-electrode setup is in principle favorable in terms of fabrication and electronic setup simplicity, and this is where we started from. Following the work of other groups [44, 45] we initially tried to compensate device stability with a better control of electrode functionalization. However, our devices still suffered from a drift in time that lasted several hours. That is why we decided to introduce a third electrode, as in more standard electrochemical setup. We believe that the parallel study of the performance of the two devices proposed in this thesis could be of great help for researchers who would like to begin to work in this field. Then, exploiting the stability of the three-electrode setup, we measured a calibration curve of our device with respect to the concentration of the complementary DNA in solution (Figure 4.17). Using this graph we could build the plot shown in Figure 4.18 and, from it, compute the hybridization affinity constant of our immobilized, thiolated DNA-molecules. The computed values are in good agreement with existing data obtained using SPR [69, 68]. Although the final goal of our work is protein detection, the measurement of DNA hybridization can be considered as a proof of principle of soundness of our setup, and is instrumental for the next functionalization step, which is based on DNA-directed protein immobilization. Finally, thanks to the stability obtained with our three-electrode configuration and due to the simplicity of the measurements environment (no redox couples), we were able to measure DNA hybridization kinetics in real time and, most important, in-situ. According to our knowledge, this is the first time that the variation of the differential capacitance is used for real time kinetic measurements. Previous EIS experiments were in fact based on the detection of charge transfer resistance variation [67], through a setup that, because of severe instabilities, obliged to perform experiments ex-situ. The possibility to study the kinetics of DNA-hybridization in real time is very valuable for biosensor design and optimization and for the efficient collection of genomic data [70]. Biologically relevant phenomena as hairpin folding, which 4.7 Major Achievements of the Ph.D. Work 69 plays a key role in biological functions as regulator of gene expression [71], or DNA single base pair flipping, fundamental in the interaction between DNA and DNA-repairing enzymes [71], can be in fact discriminated through kinetic and dynamic studies of DNA-hybridization. 5 Conclusion In this final chapter we draw the conclusions of the Ph.D.-work. In the first section we present all the achievements whereas, in the second, we point out the questions which are still open suggesting several interesting perspectives. 5.1 Where we are As mentioned in the introduction, the aim of this Ph.D.-work was to explore electrical readout systems for the detection of bio-markers useful for medical diagnostics. After a thoroughly examination of the literature, we directed our attention towards systems based on the monitoring of the double layer capacitance, C DL , at the interface electrode/electrolyte. The main reasons that influenced this decision are the potential to integrate the final device in a complete miniaturized system, the possibility of multiplexing and the chance to exploit the expertise of the group in the functionalization of gold surfaces and protein immobilization via DNA directed immobilization (DDI) [8, 52]. During the last 3 years we thus developed two complete systems based on electrochemical impedance spectroscopy (EIS). One setup was based on a microfluidic channel and two microelectrodes; the other one was composed by a classical three-electrode configuration where two electrodes, working and counter-electrode, are in the micrometer range while the reference electrode is a pellet Ag/AgCl electrode with a diameter, d = 4mm. For both configurations a specific sample-holder was designed and fabricated (see Figures 2.7 and 2.8). Proper electrical connections and shielding were produced as well. After fabrication both setups were characterized vs frequency in order to find the optimal experimental parameters for C DL -measurement. After calibration we challenged the two configurations with the detection of biological reactions: DNA-hybridization and protein-protein binding (only for the two-electrode devices). In the next paragraphs we summarize the main results. DNA-hybridization was detected using both the two- and the three-electrode configuration (see Figures 3.4 and 4.4). The two-electrode setup however, turned out to suffer from a very strong time drift, that complicates the detection and hinders the application of the device as a fast point-of-care diagnostic tool. Following the literature, we tried to passivate the electrode in order to limit and eventually avoid the capacitance drift. We explored several passivation layers: C14, MCH, mix of ssDNA and MCH. Even though the passivation of the gold electrodes could effectively reduce the drift in the capacitance-value (see Figure 3.6), it could not completely prevent it. For this reason we started to follow the capacitance 72 5 Conclusion value over time. Using this strategy we further stressed the detector varying the passivation layer and the buffer solution (see Figure 3.7 and 3.8). In this way we demonstrated that the two-electrode configuration can discriminate among different gold functionalizations but is also affected by the buffer ions in solution. In order to avoid misleading results we thus decided to keep constant the buffer throughout the experiments and test the detector upon protein-protein binding. We detected protein-immobilization via DDI using engineered streptavidin-molecules and we further detected a second immobilization step of biotinylated aGFAP-molecules (see Figure 3.11). Eventually we compared our results with literature and we found good agreement. Although we were able to detect protein-protein interactions, the two-electrode devices suffered, from time to time, from big instabilities in the capacitance signal (see Figure 3.14). Thus we decided to upgrade the setup introducing a third reference electrode. The RE should control the applied potential between WE and solution and avoid the time drift of the capacitance signal and the measured instabilities. After characterization of the RE we tested the three-electrode device upon DNA-hybridization. First we measured the capacitance at the WE as a function of the DC applied potential (see Figure 4.4). During these measurements we did not observe the minimum in the capacitance predicted for a bare electrode and we argued that the electrode was correctly passivated. Another proof of the presence of the DNA-layer on the gold surface was given by the AFM-measurements. Thanks to shaving-experiments in fact, we demonstrated directly both the presence of the functionalization layer and the DNA-hybridization (see Figure 4.6 and 4.7). After these first experiments we checked the system as a function of time. We measured the value of Cd over many hours (up to 11h) and for several passivation layers. The variations of the capacitance in time turned out to be very small and, in general, amounted to less than 1% of the initial Cd -value (see Figure 4.10). Most interestingly, the decay of Cd in time, always observed in the two-electrode devices, disappeared. At this point we started to test again the system upon DNA-hybridization over time. We introduced a plasma cleaning procedure as a consequence of the worsening of the gold quality and we demonstrated detection of DNA-hybridization in KCl 10 mM, the same buffer solution used in the experiments with the two-electrode devices (see Figure 4.13). Increasing the salt concentration to KCl 100 mM, we noticed that the MCH-molecules in the SAM prevents the detection of the DNA-hybridization. For this reason we avoided MCH post treatment and indeed we measured a big variation of the Cd -signal upon DNA-pairing (see Figure 4.15). Still avoiding the use of MCH we performed a calibration curve of the device and we found out that the limit of detection for the device corresponds to a concentration of the complementary DNA, ccDNA = 100 pM (see Section 4.4). Eventually we exploited the time stability of the three-electrode devices in order to measure DNA-hybridization in real time (see Figure 4.19). We fitted the data using Langmuir kinetics and we found out that the system can be satisfactorily modeled with a Langmuir kinetics both of the first and second order. The latter, however, fits better the experimental data at the beginning and in the end of the experiment. For this reason we believe that the conditions in which we performed our experiment are better modeled considering diffusion of the DNA-molecules towards the electrode and thus by a Langmuir kinetics of the second order. Detection of DNA-hybridization was proved even for a mix SAM composed by ssDNA and the biorepellent oligoethylene glicol molecules (TOEG6). This result could be of great importance for the future development of the sensor and the biomolecular detection in complex matrices like urine or blood-plasma. 5.2 Perspectives 73 5.2 Perspectives As mentioned in the above section, multiplexing is the final goal of the project. After the very important achievement of time stability, which we obtained with the three-electrode configuration, we plan now to further upgrade the setup to allow for multiplexing. The technologies we used, lithography and EIS, permit parallel fabrication and measurement while the experience we gathered during the Ph.D.-project will speed up the work. First of all we plan to realize a detector for DNA-hybridization for three different sequences of DNA to be analyzed in parallel. The scheme we are going to follow is the realization, on the same glass slide, of three couples of WE + CE with the same layout presented in Figure 2.4. The RE will be a common Ag/AgCl electrode positioned just above the glass slide. The latter must be removable in order to perform the functionalization of the electrodes (as explained in 2.5) and to change the experimental solutions. In a further step we plan to integrate the whole system into a microfluidic channel. In order to do it we will build free standing membranes of PDMS, where we can manually cut a microchannel. The technique to fabricate such membranes of PDMS is quite simple and consist of three steps: first one spin-coats a glass slide with a water soluble resist (e.g. polyvinyl alcohol (PVA)); second the slide is spincoated again with a layer of PDMS; third the membrane is released from the slide via lift off in water. After these steps one can manually cut a slice of the PDMS in order to shape the microchannel. We have already made some tests of this kind and we are developing the Ag/AgCl lamina, which should form the ceiling of the channel. A pictorial view of the different parts of the final device are shown in Figure 5.1. Fig. 5.1. Pictorial view of the three main parts of the device for parallel detection of different sequences of DNA within a microchannel. a) base plate constituted by a glass slide with the three couples of WE and CE. b) PDMS membrane with microchannel. c) Ag/Ag/Cl lamina which serves as RE and upper wall of the channel. Holes are drilled in the lamina in order to connect the microchannel with the pump-system. Using the explained setup, we are relatively sure to produce a microfluidic channel with incorporated electrodes with a height, h ≈ 30 µm. The problem with this device would be the targeted functionalization of the different WEs. For this aim we plan to follow the idea of Ahmad et al. reported in 2011 for the relization of a microfluidic platform for bio-markers recognition [78]. In their work Ahmad et al. show 74 5 Conclusion that it is possibile to selectively functionalize small areas of a glass slide using a network of removable channels. Following this idea we plan to create three parallel microchannels using soft lithography (see 2.2), flux within them three different solutions containing three different thiolated DNA sequences and then remove the PDMS with the three channels and apply the membrane in order to perform the detection inside the microchannel. Besides multiplexing, in the next future we further plan to continue the measurements on DNAhybridization in real time. We believe in fact, that these data could shed light on the way the kinetics of DNA-pairing takes place. Carefully fitting the experimental data obtained for different concentrations of complementary DNA in solution and in correspondence of ssDNA-SAM of different densities in fact, we could understand at which concentration the process possesses the features of a pure first order Langmuir kinetics and thus support or deny the hypothesis present in literature. Finally we would like to briefly present another idea that we are going to explore: detection of biomolecules within nanochannels by measuring the surface conductance of the channels. This idea is different from the topic of the Ph.D. in the sense that it will neither involve the fabrication techniques presented in the thesis nor the functionalization of gold electrodes with thiolated molecules. Nevertheless we will use the wide experience we have gathered in the field of electrical measurements in solution and in the realization of experimental setup. The idea is to produce a silicon membrane with a certain number (hundreds) of nanochannels (diameter d ≈ 200 nm), functionalize their internal surfaces with silane molecules and eventually measure the ionic conductance across two reservoirs divided by the membrane. This idea has been proposed in several papers but it is still very new and intriguing. The conductance characteristics of such nanochannels are still under investigation and seems to hold great perspectives [79, 80, 81, 82]. In our work we would focus on channels with an internal diameter of hundreds of nm for which clogging should not represent a problem. At the moment we are realizing the first membranes and then we will continue with conductance measurements in order to characterize them electrically. After 4 years in this field we believe that, in order to produce a successful detectors for medical diagnostics, simplicity must be the ultimate goal. In the devices we explored during the Ph.D. problems arose from almost all the aspects which we believed granted: gold purity, stability of the applied potential, cleanliness of the environment etc. In the proposed device all the fabrication materials would be of industrial purity while the measurements and the setup can be performed easily without the use of microchannels and DC-applied potentials (a problem in solution). We will not quit the initial idea of integrating microfluidic network and electrical measurements but we believe that, in order to avoid very big hurdles, the two aspects of the final device should be implemented separately and then joined only in the final stages, when the big criticalities of both parts will be solved. A Differential Capacitance at the Electrode-Electrolyte Interface: Models of the Differential Capacitance In the Introduction 1 we mentioned that the interface between electrode and electrolyte is the most interesting part of the electrochemical circuit shown in Figure 1.1. In 1853 Helmholtz tried to model it for the first time and had a simple but very intelligent idea. Knowing that, at equilibrium, the charge in the metallic electrode is distributed at its surface, he imagined that the ions in solution behave in the same way and distribute on a surface at a distance d from the electrode. The ions counterbalance exactly the charge on the metal and form, together with the electrode, the double layer capacitance, C DL . This simple model implies that, given a potential V between the two surfaces, the charge density, σ, stored by C DL becomes: 0 V (A.1) d where and 0 represent the dielectric constant of solution and vacuuum, respectively. From Equation σ= A.1 we can further compute the corresponding differential capacitance, Cd , as: ∂σ 0 = . (A.2) ∂V d From Equation A.2 we recognize immediately the weakness of the model proposed by Helmholtz, Cd = i.e., that Cd is a constant independent of the applied voltage. This feature disagrees with the experiments and thus more sophisticated models to describe the interface electrode/electrolyte are necessary. One very good approximation is the model proposed by Gouy and Chapman and improved by Stern in 1924 with the introduction of a layer of adsorbed ions on the surface. According to this model the ions in solutions can either stick on the surface or stay in the so called diffusive layer, where the ions reorganize according to Boltzmann’s distribution. Conventionally one defines two planes: the inner Helmholtz plane (IHP), the plane that passes along the centers of the attached atoms, and the outer Helmholtz plane (OHP), the plane which intersects the centers of the hydrated atoms at the minimum distance from the surface. In Figure A.1 we show a pictorial image that describes the system. Following this idea one can compute the charge density on the metal electrode, σ M : σM = p 8kB T 0 n0 sinh[ Ze σ M xIHP (Φ0 − )] 2kB T 0 (A.3) 78 A Differential Capacitance at the Electrode-Electrolyte Interface: Models of the Differential Capacitance Fig. A.1. Model of the electrode/electrolyte interface. The ions in solutions can either stick on the surface, within the inner Helmholtz plane or remain in the so called diffusive layer, beyond the outer Helmholtz plane. In the diffusive layer the ions reorganize according to Boltzmann’s distribution. where kB is the Boltzmann constant, T the absolute temperature, n0 the ions density in the bulk solution, far from the surface, Z the valence of the ions, Φ0 the potential at the metallic surface, xIHP the distance of the IHP from the surface. Differentiating Equation A.3 one obtain the following equation that describes the inverse of the differential capacitance, Cd , at the electrode/electrolyte interface: 1 ∂σ M − xOHP 1 =( ) 1= + p 2 2 0 Cd ∂Φ0 0 20 Z e n /kB T cosh(ZeΦOHP /2kB T ) (A.4) In Figure A.2 we further show the potential profile, φ, as a function of the distance from the electrode, x. From the capacitance point of view, Cd is composed by two capacitances in series, the one coming from the ions attached to the metallic surface and the one due to the ions diffused in solution. As a function of the applied potential one can tune the contribution of the two capacitances and highlight either the first one or the second one. In Figure A.3 we plot the theoretical behavior of Cd as a function of the applied voltage E. In the graph we note that, for very positive and negative voltages the Stern’s layer dominate and the capacitance is constant. At potentials equal to the point of zero charge, the potential for which the attached ions at the surface counterbalance the charge density of the electrode, the capacitance has a parabolic shape generated by the diffused ions. In real examples the situation is obviously a bit different and the theoretical behavior can diverge from the experimental points. Nevertheless this model is very useful and helps in understanding the behavior of the differential capacitance at the electrode/electrolyte interface. A Differential Capacitance at the Electrode-Electrolyte Interface: Models of the Differential Capacitance 79 Fig. A.2. Potential profile, φ, as a function of the distance from the electrode, x. The distance x2 corresponds to the IHP. Fig. A.3. Theoretical behavior of Cd as a function of the applied voltage E at the electrode. For very positive and negative voltages the Stern’s layer dominate and the capacitance is constant. At potentials equal to the point of zero charge, the potential for which the attached ions at the surface counterbalance the charge density of the electrode, the capacitance has a parabolic shape generated by the diffused ions 80 A Differential Capacitance at the Electrode-Electrolyte Interface: Models of the Differential Capacitance The treatment of the differential capacitance proposed in this chapter is intended to be a brief support during the lecture of the thesis. All the figures and the equations presented in this appendix have been extracted from the book <<Electrochemical Methods: Fundamentals and Applications>> [65]. A more detailed treatment can be found therein. B Instruments and Lithographic Procedures During the last four years I acquired a remarkable experience in the field of microfabrication. In this appendix I list the main instruments we used for the fabrication of the devices: microelectrodes and microchannels. B.1 Spin-coater In Figure B.1 we show a picture of the spin-coater, model CPS20, we used for spinning the resist, both positive and negative, on the substrate to be processed: glass and silicon. Fig. B.1. Picture of the spin-coater used during the thesis-work. Its main features are listed in Table B.1. The main features of the spin-coater are listed in Table B.1. 82 B Instruments and Lithographic Procedures Model CPS20 Operation mode automatic and programmable Max number of programs 6 Max spin-speed (9000 ± 10) rpm Min spin-speed (100 ± 10) rpm Table B.1. Main features of the spin-coater CPS20. B.2 Mask Aligners 83 B.2 Mask Aligners In order to perform photo-lithography we made use of two distinct mask-aligners: the semi-automatic mask-aligner MA25 and the manual one MJB3. Both instruments were produced by the company Karl Suss, Germany. In Figure B.2 and B.3 we show both instruments whereas in Table B.2 we list their main characteristics. Fig. B.2. Picture of the mask aligner MJB3 used during the thesis-work. Its main features are listed in Table B.2. Fig. B.3. Picture of the mask aligner MA25. Its main features are listed in Table B.2. 84 B Instruments and Lithographic Procedures Model MJB3 MA25 Producer Karl Suss Karl Suss Operation mode manual semi-automatic UV-lamp Mercury Mercury Max wafer size 4 inch 6 inch Double exposure No Yes Alignment of Yes limited 1µm 1µm mask and sample Minimal size of achievable features Table B.2. Main features of the mask aligners used during the Ph.D. B.3 e-beam Evaporator 85 B.3 e-beam Evaporator In order to fabricate the micro-electrodes we employed an electron-beam evaporator: Rial EGE 450. Thanks to this machine we were able to deposit thin films of gold and titanium with a precision of few nm’s. In Figure B.4 we show a picture of the instrument and in Table B.3 its main features. Fig. B.4. Picture of the e-beam evaporator used during the Ph.D. Its main features are listed in Table B.3. Model Rial EGE 450 Operation mode automatic number of crucibles 4 Max wafer size 4 inch distance crucible/sample 40 cm Operation pressure ≈ 1 · 10−6 mbar Operation temperature ≈ 18 K Thickness control Quartz micro-balance Table B.3. Main features of the e-beam evaporator Rial EGE 450. 86 B Instruments and Lithographic Procedures B.4 Lithografic Procedures During the Ph.D. we employed different lithographic procedures according to the different resists, negative or positive, and to the desired features of the final devices. In Table B.4 we report all the steps that one has to carry out in order to perform lithography with S1818 and SU8-100, the two main resist used during our work. S1818 (positive resist) was used for the shaping of the electrodes and for final isolation of the three-electrode devices whereas SU8-100 (negative resist) was used for the production of the mold for the fabrication of the micro-channels. With the parameters listed in Table B.4 one obtains a resist thickness of roughly 2.5 µm and 65 µm for S1818 and SU8-100, respectively. Resist S18-18 Dehydration of the support (Si or Glass) hot plate 200 °C for 5 min Spinning of Omnicoat SU8-100 hot plate 200 °C for 5 min No 500 rpm for 5 s and 3000 rpm for 20 s Bake of Omnicoat No hot plate 200 °C for 1 min Spinning 3000 rpm for 60 s 1000 rpm for 5 min Pre-bake hot plate 115 °C for 1 min hot plate 65 °C for 30 min 95 °C for 50 min Temperature Ramps 10 min Exposure 21 s @ 3 mW/cm2 40 s @ 3 mW/cm2 Post-bake No hot plate 65 °C for 2 min 95 °C for 20 min Temperature Ramps 10 min Development ≈ 30 s in MF319 ≈ 2 min in SU8-Developer Rinse in purified water in Isopropanol Table B.4. Lithographic steps to be performed for a positive and a negative resist, S1818 and SU8-100, respectively. Prior to SU8-spinning one has to spin-coat the sample with Omnicoat. This layer is necessary in order to guarantee better adhesion of the SU8 with the substrate. References 1. Vladimir Gubala, Leanne F Harris, Antonio J Ricco, Ming X Tan, and David E Williams. Point of care diagnostics: status and future. Analytical chemistry, 84(2):487–515, 2011. 2. V Tsouti, C Boutopoulos, I Zergioti, and S Chatzandroulis. Capacitive microsystems for biological sensing. Biosensors and Bioelectronics, 27(1):1–11, 2011. 3. Sandro Carrara. Nano-Bio-Technology and Sensing Chips: New Systems for Detection in Personalized Therapies and Cell Biology. Sensors, 10(1):526–543, January 2010. 4. Shaurya Prakash, M.B. Karacor, and S. Banerjee. Surface modification in microsystems and nanosystems. Surface Science Reports, 64(7):233–254, July 2009. 5. Paul Yager, Gonzalo J Domingo, and John Gerdes. Point-of-care diagnostics for global health. Annual review of biomedical engineering, 10:107–44, January 2008. 6. Xiaole Mao and Tony Jun Huang. Microfluidic diagnostics for the developing world. Lab on a chip, 12(8):1412– 6, April 2012. 7. Bong-Hyun Jun, Homan Kang, Yoon-Sik Lee, and Dae Hong Jeong. Fluorescence-based multiplex protein detection using optically encoded microbeads. Molecules (Basel, Switzerland), 17(3):2474–90, January 2012. 8. Fouzia Bano, Ljiljana Fruk, Barbara Sanavio, Maximilian Glettenberg, Loredana Casalis, Christof M Niemeyer, and Giacinto Scoles. Toward multiprotein nanoarrays using nanografting and dna directed immobilization of proteins. Nano letters, 9(7):2614–2618, 2009. 9. Barbara Sanavio, Denis Scaini, Christian Grunwald, Giuseppe Legname, Giacinto Scoles, and Loredana Casalis. Oriented immobilization of prion protein demonstrated via precise interfacial nanostructure measurements. Acs Nano, 4(11):6607–6616, 2010. 10. Marilina Tampoia, Davide Giavarina, Chiara Di Giorgio, and Nicola Bizzaro. Diagnostic accuracy of enzymelinked immunosorbent assays (ELISA) to detect anti-skin autoantibodies in autoimmune blistering skin diseases: a systematic review and meta-analysis. Autoimmunity reviews, 12(2):121–6, December 2012. 11. Helen Ludlow, David J Phillips, Michelle Myers, Robert I McLachlan, David M de Kretser, Carolyn a Allan, Richard a Anderson, Nigel P Groome, Marko Hyvönen, W Colin Duncan, and Shanthi Muttukrishna. A new ’total’ activin B enzyme-linked immunosorbent assay (ELISA): development and validation for human samples. Clinical endocrinology, 71(6):867–73, December 2009. 12. a Venteo, B Rebollo, J Sarraseca, M J Rodriguez, and a Sanz. A novel double recognition enzyme-linked immunosorbent assay based on the nucleocapsid protein for early detection of European porcine reproductive and respiratory syndrome virus infection. Journal of virological methods, 181(1):109–13, April 2012. 13. Christina Boozer, Jon Ladd, Shengfu Chen, Qiuming Yu, Jiri Homola, and Shaoyi Jiang. Dna directed protein immobilization on mixed ssdna/oligo (ethylene glycol) self-assembled monolayers for sensitive biosensors. Analytical Chemistry, 76(23):6967–6972, 2004. 88 References 14. Sanna Auer, Martin Nirschl, Matthias Schreiter, and Inger Vikholm-Lundin. Detection of DNA hybridisation in a diluted serum matrix by surface plasmon resonance and film bulk acoustic resonators. Analytical and bioanalytical chemistry, 400(5):1387–96, May 2011. 15. Luca Ferrari, Hana Šı́pová, Ivo Tichý, Karel Chadt, and Jiri Homola. Electrochemical surface plasmon resonance biosensor for study of DNA desorption and hybridization. Optical Sensors, 8774:87740F–87740F–9, May 2013. 16. a. Toma, G. Das, M. Chirumamilla, a. Saeed, R. Proietti Zaccaria, L. Razzari, M. Leoncini, C. Liberale, F. De Angelis, and E. Di Fabrizio. Fabrication and characterization of a nanoantenna-based Raman device for ultrasensitive spectroscopic applications. Microelectronic Engineering, 98:424–427, October 2012. 17. Justin L Abell, Jeonifer M Garren, Jeremy D Driskell, Ralph a Tripp, and Yiping Zhao. Label-Free Detection of Micro-RNA Hybridization Using Surface-Enhanced Raman Spectroscopy and Least-Squares Analysis. Journal of the American Chemical Society, pages 1–4, July 2012. 18. E Gosselin, M Gorez, M Voué, O Denis, J Conti, N Popovic, a Van Cauwenberge, E Noel, and J De Coninck. Fourier transform infrared immunosensors for model hapten molecules. Biosensors & bioelectronics, 24(8):2554–8, April 2009. 19. Per Rigler, Wolf-Peter Ulrich, Patrik Hoffmann, Michael Mayer, and Horst Vogel. Reversible immobilization of peptides: surface modification and in situ detection by attenuated total reflection FTIR spectroscopy. Chemphyschem : a European journal of chemical physics and physical chemistry, 4(3):268–75, March 2003. 20. Zhong Zhang, Mengshi Lin, Sha Zhang, and Bongkosh Vardhanabhuti. Detection of aflatoxin m1 in milk by dynamic light scattering coupled with superparamagnetic beads and gold nanoprobes. Journal of agricultural and food chemistry, 2013. 21. J L Arlett, E B Myers, and M L Roukes. Comparative advantages of mechanical biosensors. Nature nanotechnology, 6(4):203–15, April 2011. 22. Mauro Melli, Giacinto Scoles, and Marco Lazzarino. Fast detection of biomolecules in diffusion-limited regime using micromechanical pillars. ACS nano, 5(10):7928–35, October 2011. 23. Min Yue, Jeanne C Stachowiak, Henry Lin, Ram Datar, Richard Cote, and Arun Majumdar. Label-free protein recognition two-dimensional array using nanomechanical sensors. Nano letters, 8(2):520–4, February 2008. 24. Timothy R J Holford, Frank Davis, and Séamus P J Higson. Recent trends in antibody based sensors. Biosensors & bioelectronics, 34(1):12–24, April 2012. 25. Wei-An Lai, Chih-Heng Lin, Yuh-Shyong Yang, and Michael S-C Lu. Ultrasensitive and label-free detection of pathogenic avian influenza DNA by using CMOS impedimetric sensors. Biosensors & bioelectronics, 35(1):456– 60, May 2012. 26. Kang-Ho Lee, Jeong-Oen Lee, Mi-Jin Sohn, Byunghun Lee, Suk-Hwan Choi, Sang Kyu Kim, Jun-Bo Yoon, and Gyu-Hyeong Cho. One-chip electronic detection of DNA hybridization using precision impedance-based CMOS array sensor. Biosensors & bioelectronics, 26(4):1373–9, December 2010. 27. I-Jane Chen and Ian M White. High-sensitivity electrochemical enzyme-linked assay on a microfluidic interdigitated microelectrode. Biosensors & bioelectronics, 26(11):4375–81, July 2011. 28. M. Javanmard, H. Esfandyarpour, F. Pease, and R. W. Davis. Electrical detection of proteins and DNA using bioactivated microfluidic channels: Theoretical and experimental considerations. Journal of Vacuum Science & Technology B: Microelectronics and Nanometer Structures, 27(6):3099, 2009. 29. Mehdi Javanmard, Amirali H Talasaz, Mohsen Nemat-Gorgani, Fabian Pease, Mostafa Ronaghi, and Ronald W Davis. Electrical detection of protein biomarkers using bioactivated microfluidic channels. Lab on a chip, 9(10):1429–34, May 2009. 30. M. Javanmard and R.W. Davis. A microfluidic platform for electrical detection of DNA hybridization. Sensors and Actuators B: Chemical, March 2010. References 89 31. D. C. Martins, V. Chu, D. M. F. Prazeres, and J. P. Conde. Electrical detection of DNA immobilization and hybridization by streaming current measurements in microchannels. Applied Physics Letters, 99(18):183702, 2011. 32. Michelle L Kovarik and Stephen C Jacobson. Nanofluidics in lab-on-a-chip devices. Analytical chemistry, 81(17):7133–40, September 2009. 33. Jongin Hong, Dae Sung Yoon, Sung Kwan Kim, Tae Song Kim, Sanghyo Kim, Eugene Y Pak, and Kwangsoo No. AC frequency characteristics of coplanar impedance sensors as design parameters. Lab on a chip, 5(3):270–9, March 2005. 34. Christine Berggren, Bjarni Bjarnason, and Gillis Johansson. Capacitive Biosensors. Electroanalysis, 13(3):173– 180, March 2001. 35. Jonathan S Daniels and Nader Pourmand. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis, 19(12):1239–1257, May 2007. 36. Rohit Karnik, Chuanhua Duan, Kenneth Castelino, Hirofumi Daiguji, and Arun Majumdar. Rectification of ionic current in a nanofluidic diode. Nano letters, 7(3):547–51, March 2007. 37. Nicolas F Y Durand and Philippe Renaud. Label-free determination of protein-surface interaction kinetics by ionic conductance inside a nanochannel. Lab on a chip, 9(2):319–24, January 2009. 38. Ivan Vlassiouk, Thomas R Kozel, and Zuzanna S Siwy. Biosensing with nanofluidic diodes. Journal of the American Chemical Society, 131(23):8211–20, June 2009. 39. Rohit Karnik, Kenneth Castelino, Rong Fan, Peidong Yang, and Arun Majumdar. Effects of biological reactions and modifications on conductance of nanofluidic channels. Nano letters, 5(9):1638–42, September 2005. 40. Peter Van Gerwen, Wim Laureyn, Wim Laureys, Guido Huyberechts, Maaike Op De Beeck, Kris Baert, Jan Suls, Willy Sansen, P Jacobs, Lou Hermans, et al. Nanoscaled interdigitated electrode arrays for biochemical sensors. Sensors and Actuators B: Chemical, 49(1):73–80, 1998. 41. a Martinez-Rivas, F Carcenac, D Saya, C Séverac, L Nicu, and C Vieu. Wafer scale interdigitated nanoelectrode devices functionalized using a MEMS-based deposition system. Nanotechnology, 23(10):105302, March 2012. 42. Liju Yang, Yanbin Li, Carl L Griffis, and Michael G Johnson. Interdigitated microelectrode (IME) impedance sensor for the detection of viable Salmonella typhimurium. Biosensors & bioelectronics, 19(10):1139–47, May 2004. 43. Zhiwei Zou, Junhai Kai, Michael J. Rust, Jungyoup Han, and Chong H. Ahn. Functionalized nano interdigitated electrodes arrays on polymer with integrated microfluidics for direct bio-affinity sensing using impedimetric measurement. Sensors and Actuators A: Physical, 136(2):518–526, May 2007. 44. Sandro Carrara, Vijayender Bhalla, Claudio Stagni, and Bruno Samorı̀. Nanoscale film structure related to capacitive effects in ethylene-glycol monolayers. Surface Science, 603(13):L75–L77, July 2009. 45. V M Mirsky, M Riepl, and O S Wolfbeis. Capacitive monitoring of protein immobilization and antigen-antibody reactions on monomolecular alkylthiol films on gold electrodes. Biosensors & bioelectronics, 12(9-10):977–89, January 1997. 46. Jianhua Zhou, Kangning Ren, Yizhe Zheng, Jing Su, Yihua Zhao, Declan Ryan, and Hongkai Wu. Fabrication of a microfluidic Ag/AgCl reference electrode and its application for portable and disposable electrochemical microchips. Electrophoresis, 31(18):3083–9, September 2010. 47. M Waleed Shinwari, David Zhitomirsky, Imran a Deen, P R Selvaganapathy, M Jamal Deen, and D Landheer. Microfabricated reference electrodes and their biosensing applications. Sensors (Basel, Switzerland), 10(3):1679– 715, January 2010. 48. Brian J. Polk, Anna Stelzenmuller, Geraldine Mijares, William MacCrehan, and Michael Gaitan. Ag/AgCl microelectrodes with improved stability for microfluidics. Sensors and Actuators B: Chemical, 114(1):239–247, March 2006. 90 References 49. C Guiducci, C Stagni, G Zuccheri, A Bogliolo, L Benini, B Samor, and B Ricc. A biosensor for direct detection of dna sequences based on capacitance measurements. In Proc. of the 32nd European Solid-State Device Research Conference, pages 479–482, 2002. 50. C Guiducci. DNA detection by integrable electronics. Biosensors and Bioelectronics, 19(8):781–787, March 2004. 51. a W Peterson, R J Heaton, and R M Georgiadis. The effect of surface probe density on DNA hybridization. Nucleic acids research, 29(24):5163–8, December 2001. 52. Elham Mirmomtaz, Matteo Castronovo, Christian Grunwald, Fouzia Bano, Denis Scaini, Ali a Ensafi, Giacinto Scoles, and Loredana Casalis. Quantitative study of the effect of coverage on the hybridization efficiency of surface-bound DNA nanostructures. Nano letters, 8(12):4134–9, December 2008. 53. Rastislav Levicky, Tonya M. Herne, Michael J. Tarlov, and Sushil K. Satija. Using Self-Assembly To Control the Structure of DNA Monolayers on Gold: A Neutron Reflectivity Study. Journal of the American Chemical Society, 120(38):9787–9792, September 1998. 54. Frank Schreiber. Structure and growth of self-assembling monolayers. Progress in surface science, 65(5):151– 257, 2000. 55. M. Lazerges, H. Perrot, N. Rabehagasoa, C. Compère, C. Dreanno, M. Mucio Pedroso, R.C. Faria, and P.R. Bueno. DNA hybridization mechanism in an interfacial environment: What hides beneath first order k (s1) kinetic constant? Sensors and Actuators B: Chemical, 171-172:522–527, August 2012. 56. Larry J Kricka. Microchips, microarrays, biochips and nanochips: personal laboratories for the 21st century. Clinica Chimica Acta, 307(1):219–223, 2001. 57. Christine Berggren, Per Stålhandske, Jan Brundell, and Gillis Johansson. A feasibility study of a capacitive biosensor for direct detection of dna hybridization. Electroanalysis, 11(3):156–160, 1999. 58. Sandro Carrara, Andrea Cavallini, Yusuf Leblebici, Giovanni De Micheli, Vijayender Bhalla, Francesco Valle, Bruno Samorı̀, Luca Benini, Bruno Riccò, and Inger Vikholm-Lundin. Capacitance DNA bio-chips improved by new probe immobilization strategies. Microelectronics Journal, 41(11):711–717, November 2010. 59. Archana N Rao, Christopher K Rodesch, and David W Grainger. Real-time fluorescent image analysis of DNA spot hybridization kinetics to assess microarray spot heterogeneity. Analytical chemistry, 84(21):9379– 87, November 2012. 60. Sandro Carrara, Vijayender Bhalla, Claudio Stagni, Luca Benini, Anna Ferretti, Francesco Valle, Andrea Gallotta, Bruno Riccò, and Bruno Samorı̀. Label-free cancer markers detection by capacitance biochip. Sensors and Actuators B: Chemical, 136(1):163–172, February 2009. 61. Sandro Carrara, Luca Benini, Vijayender Bhalla, Claudio Stagni, Anna Ferretti, Andrea Cavallini, Bruno Riccò, and Bruno Samorı̀. New insights for using self-assembly materials to improve the detection stability in label-free DNA-chip and immuno-sensors. Biosensors & bioelectronics, 24(12):3425–9, August 2009. 62. M L Foresti, F Loglio, M Innocenti, S Bellassai, F Carlà, E Lastraioli, G Pezzatini, C Bianchini, and F Vizza. Confined electrodeposition of CdS in the holes left by the selective desorption of 3-mercapto-1-propionic acid from a binary self-assembled monolayer formed with 1-octanethiol. Langmuir : the ACS journal of surfaces and colloids, 26(3):1802–6, February 2010. 63. Daisuke Oyamatsu, Takeshi Fujita, Satoshi Arimoto, Hirokazu Munakata, Hajime Matsumoto, and Susumu Kuwabata. Electrochemical desorption of a self-assembled monolayer of alkanethiol in ionic liquids. Journal of Electroanalytical Chemistry, 615(2):110–116, April 2008. 64. Cindra A Widrig, Chinkap Chung, and Marc D Porter. The electrochemical desorption of n-alkanethiol monolayers from polycrystalline au and ag electrodes. Journal of electroanalytical chemistry and interfacial electrochemistry, 310(1):335–359, 1991. References 91 65. Allen J Bard and Larry R Faulkner. Electrochemical methods: fundamentals and applications, volume 2. Wiley New York, 1980. 66. Attila Bonyar and Gábor Harsanyi. Afm nanoshaving: A novel prospect for the structural comparison of bioreceptor layers. In Electronics Technology (ISSE), 2011 34th International Spring Seminar on, pages 519–524. IEEE, 2011. 67. D Li, X Zou, Q Shen, and S Dong. Kinetic study of DNA/DNA hybridization with electrochemical impedance spectroscopy. Electrochemistry Communications, 9(2):191–196, February 2007. 68. B Persson, K Stenhag, P Nilsson, a Larsson, M Uhlén, and P Nygren. Analysis of oligonucleotide probe affinities using surface plasmon resonance: a means for mutational scanning. Analytical biochemistry, 246(1):34–44, March 1997. 69. D Kambhampati, P E Nielsen, and W Knoll. Investigating the kinetics of DNA-DNA and PNA-DNA interactions using surface plasmon resonance-enhanced fluorescence spectroscopy. Biosensors & bioelectronics, 16(912):1109–18, December 2001. 70. Feng Long, Shuxu Wu, Miao He, Tiezheng Tong, and Hanchang Shi. Ultrasensitive quantum dots-based DNA detection and hybridization kinetics analysis with evanescent wave biosensing platform. Biosensors & bioelectronics, 26(5):2390–5, January 2011. 71. Yandong Yin and Xin Sheng Zhao. Kinetics and dynamics of dna hybridization. Accounts of Chemical Research, 44(11):1172–1181, 2011. 72. Yu Liu and Liang Shen. From Langmuir kinetics to first- and second-order rate equations for adsorption. Langmuir : the ACS journal of surfaces and colloids, 24(20):11625–30, October 2008. 73. Alexander W Peterson, Lauren K Wolf, and Rosina M Georgiadis. Hybridization of mismatched or partially matched dna at surfaces. Journal of the American Chemical Society, 124(49):14601–14607, 2002. 74. Ian Y Wong and Nicholas a Melosh. An electrostatic model for DNA surface hybridization. Biophysical journal, 98(12):2954–63, June 2010. 75. Olivier Y.F. Henry and Ciara K. OSullivan. Rapid DNA hybridization in microfluidics. TrAC Trends in Analytical Chemistry, 33:9–22, March 2012. 76. JP Tosar, G Branas, and J Laı́z. Electrochemical dna hybridization sensors applied to real and complex biological samples. Biosensors and Bioelectronics, 26(4):1205–1217, 2010. 77. P Harder, M Grunze, R Dahint, GM Whitesides, and PE Laibinis. Molecular conformation in oligo (ethylene glycol)-terminated self-assembled monolayers on gold and silver surfaces determines their ability to resist protein adsorption. The Journal of Physical Chemistry B, 102(2):426–436, 1998. 78. Habib Ahmad, Alex Sutherland, Young Shik Shin, Kiwook Hwang, Lidong Qin, Russell-John Krom, and James R Heath. A robotics platform for automated batch fabrication of high density, microfluidics-based dna microarrays, with applications to single cell, multiplex assays of secreted proteins. Review of Scientific Instruments, 82(9):094301–094301, 2011. 79. Chuanhua Duan and Arun Majumdar. Anomalous ion transport in 2-nm hydrophilic nanochannels. Nature nanotechnology, 5(12):848–52, December 2010. 80. Hirofumi Daiguji. Ion transport in nanofluidic channels. Chemical Society reviews, 39(3):901–11, March 2010. 81. Guanbin Li, Shili Wang, Chang Kyu Byun, Xiayan Wang, and Shaorong Liu. A quantitative model to evaluate the ion-enrichment and ion-depletion effect at microchannel-nanochannel junctions. Analytica chimica acta, 650(2):214–20, September 2009. 82. Derek Stein, Maarten Kruithof, and Cees Dekker. Surface-Charge-Governed Ion Transport in Nanofluidic Channels. Physical Review Letters, 93(3):1–4, July 2004. Acknowledgements I will remember the last four years for the rest of my life. I had good and bad experiences, very happy and very gloomy days (fortunately the former were much more numerous than the latter). For the first time in my life I faced problems that I could not solve on my own and the aid of other people was essential. I can say that the Ph.D. trained me and made me fit for the upcoming life. I believe that now, after 4 years of exercises, I can start a new life. I know what I would like to do and how I would like to do it. I am not a foreseer and therefore I cannot predict my future but surely I am ready. For all this I am grateful towards many people. First of all I list my friends...yes, my friends...for this time my bosses come later. In 4 years I met many wonderful people, inside and outside Sissa. I just list them as they come into my mind: Antonio, Stefania, Alessandra, Daniele, Giorgia, Gianpaolo, Giuseppe, Ina, Xiao, Loredana, Alessandro, Pietro, Pietro, Alessandro, Maryse, Elena, Jashmini, Luca, Paolo, Giacomo, Simone, Olga, Paola, Duvan, Rossana, Denis, Caterina, Amna, Davide, Daniele, Daniele, Federica, Alessandro, Simone, Gianluca, Massimo, Elisa, Gianluca, Nicola, past and present member of the Sissa football team, present and past member of the Piedi di Balsa football team and many others that surely I am forgetting. I do not want to say what I did and with whom, for they are all important. I further thank my family and all the people who work in the workshop of Elettra. What about the bosses? They are my friends and thus they are in the list, together with the others. List of Tables 2.1 General features of the purchased DNA- and thiol-molecules. The melting temperature is referred to the dsDNA. The ssDNA that binds to the gold surface is functionalized at the 5’-end with a thiol composed by a chain of 6 carbon-atoms. The sequence is referred as the sequence of the nucleotides for the DNA-molecules and the sequence of the atoms for the thiols and the top terminated oligoethylene glycol (TOEG6). . . . . . . . . . . . . . . . . . . . 11 2.2 General features of the used salt. Stock solutions of the different salts were prepared weighting the right amount of powder on a balance (accuracy 1 mg) and adding milli-Q water (resistivity ρ = 18.2 MΩcm) by means of calibrated pipettes. PBS has no specific molecular weight because it is a mixture of different salt (NaCl 137 mM, KCl 2.7 mM, Na2 HPO4 10 mM, KH2 PO4 2.0 mM). . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12 2.3 Main features of the used silicon compound: PDMS. The data have been copied from the Dow Corning website. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12 2.4 Main features of the lock-in amplifier, SR830 by SRS. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20 2.5 Main features of the bipotentiostat, PG340 usb by Heka. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22 2.6 Main features of the atomic force microscope, MFP 3D by Asylum Research. More information can be found on the Asylum Research web-page. . . . . . . . . . . . . . . . . . . . . . . . . 24 4.1 Average-values and root mean squared deviations of the values of ∆VRE−sRE computed from the OCP measurements in different salt solutions. The averaging was performed over a time interval of 1 min. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47 4.2 Fitting parameters of the gaussian equation for the shaving experiments of ssDNA- and dsDNA-SAM. In the case of dsDNA we were able to fit the experimental points with the sum of only two gaussians whereas for the ssDNA we had to use a third gaussian. The reason is the bright area in Figure 4.6 located at the border of the hole. This dirtiness is represented in the histogram and requires another gaussian for the fitting. The meaning of the parameters can be extrapolated by Equation 4.2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 53 4.3 Parameters of the plasma that we used in order to etch the first 2 nm of gold and expose in this way the clean metal. The plasma treatment was performed in the chamber of the machine for reactive ione etching (RIE). . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 57 96 List of Tables 4.4 Parameters of Equations 4.8 and 4.9 obtained through the fitting of the Cd -profile shown in Figure 4.19. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 66 B.1 Main features of the spin-coater CPS20. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 82 B.2 Main features of the mask aligners used during the Ph.D. . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84 B.3 Main features of the e-beam evaporator Rial EGE 450. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 85 B.4 Lithographic steps to be performed for a positive and a negative resist, S1818 and SU8-100, respectively. Prior to SU8-spinning one has to spin-coat the sample with Omnicoat. This layer is necessary in order to guarantee better adhesion of the SU8 with the substrate. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86 List of Figures 1.1 Representation of a two-electrode detector in a microfluidic channel. The potential is applied across the two electrodes which, on their own, are connected by the ionic solution present in the microchannel (thick black line that connects the inlet- and the outlet-tube). The interfaces electrode/electrolyte are the key-parts of the circuit, where there is a discontinuity in the nature of the charge carriers which, electrons in the lead-wires, become ions in solution. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1.2 4 Electrical model of the physical system shown in Figure 1.1. C DL models the interface electrode/electrolyte; C stray the capacitance that exists across the two electrodes while Rchannel represents the ionic resistance of the channel. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1.3 6 Scheme of the interdigitated electrodes (IDEs). (a) Image of the two electrodes. The finger-like structures of the electrode on the left intersect the same structures of the electrode on the right. (b) Zoom-in of the IDEs fingers. Each finger has a width, w, and is separated from the next one by a distance, d. Both d and w can vary from few nanometers to some micrometers. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1.4 7 Idealization of the electrode-electrolyte interface. The first layer, directly in contact with the gold surface, consists of the biological molecules and is modeled by the capacitance Cmol . The ions just above the first layer are modeled with a second capacitance, Cions , in series with the first one. Variations of Cmol arise upon molecular recognition. . . . . . . . . . . . . 2.1 8 Fabrication steps during photolithography as a function of the used resist (positive or negative). After dehydration (a), the slides are spin-coated with a photosensible resist (b) and baked on a hot plate (c). The slides are then ready for UV-exposure (d) and then, as a function of the resist, for post-bake (only negative resists) and development (e) and (f). . . . 14 2.2 Electrodes fabrication. After lithography the microscope slides are inserted in an e-beam evaporator and the metals, Ti as adhesion layer and Au, are evaporated (a,b). After metallization the metals in excess is eliminated via lift-off (c). . . . . . . . . . . . . . . . . . . . . . . . . 15 2.3 Picture of the microfabricated electrodes for the two-electrode setup. The gold electrodes were fabricated on half a microscope slide and are composed by two thin metallic films: 50 nm of gold and 50 nm of titanium. Ti acts as adhesion layer between gold and glass. . . . 15 98 List of Figures 2.4 a) Picture of the microfabricated WE and CE for the three-electrode setup. The gold electrodes were fabricated on half a microscope slide and appear black in the image. The patterned insulation layer (S1818; thickness: 2.5 µm) appears pink. b) Zoom-in of the central part of the picture. The diameter of the WE in contact with the solution is 100 µm. The patterned resist used to electrically insulate the electrodes is clearly recognizable as the darker-gray outer area of the image. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16 2.5 Fabrication steps during soft-lithography. First the PDMS is poured on the mold of SU8-100 (a) and then transferred on a hot plate for curing (b). Finally the PDMS is gently peeled-off the slides (c). . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16 2.6 PDMS block with peek orange tubes and the microfluidic channel. The channel connects the inlet and outlet tubes and is visible as a light grey line among them. . . . . . . . . . . . . . . . . 17 2.7 Sample holder for the two-electrode devices. The microscope slides with the micro-electrodes are costrained between two plexiglas slides: the micro-channel is placed orthogonally to the electrodes exposing to the solution an area of the electrodes of about 100x100 µm2 . Connection wires are directly soldered at the two gold pads (5x5 mm2 ) with an Indium drop and then connected with the instrument . . . . . . . . . . . . . . . . . . . . . . . . . 18 2.8 a) Sample holder for the three-electrode devices. The microscope slides with the micro-electrodes are costrained between two plexiglas slides. Because of the dimension of the reference electrode (diameter, d = 5 mm), the measurements were not carried out in the microfluidic channel but instead we used a small pool with a diameter of 6 mm and a height of 4 mm, holding a 100 µL volume. The reference Ag/AgCl pellet electrode is inserted directly in the solution through the hole in the top plexiglas slide and placed just above the microelectrodes. b) The electrical signal is collected from the gold pads (4x4 mm2 ) by a circuit board of custom design with SMA connectors and spring-loaded pins. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 19 2.9 a) Faraday cage designed in order to shield the external electromagnetic noise. The cage is made of Aluminium plates with a thickness of 1 cm and has a total weight of roughly 10 kg. The weight prevents mechanical vibrations at low frequencies to disturb the measurement. b) The Faraday cage permit the use of 5 BNC connection cables. The BNC connector are isolated from the cage but can be grounded through a wire (green wire in the picture). The cage itself can be grounded through a banana-connector. . . . . . . . . 19 2.10 Front panel of the lock-in amplifier, model SR830 by Stanford Research Systems. . . . . . . . 20 2.11 Steps implemented by the program written for the lock-in amplifier. After the initialization of the parameters the programs enter the main loop and leave it only when the set frequency is higher than fEND , the end frequency of the measurement. . . . . . . . . . . . 21 2.12 Picture of the transresistance amplifier DLPCA 200 by Femto. The current that enters is amplified and transformed in a proportional voltage signal. The amplifier can be used both for DC and AC current-signal and the amplification factor can be tuned in a very broad range, from 103 to 1011 V/A. According to the amplification factor the band width of the instrument decreases from 500 kHz to 1 kHz. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 21 List of Figures 99 2.13 Front panel of the bipotentiostat, model PG340 usb by Heka. . . . . . . . . . . . . . . . . . . . . . . . . . 23 2.14 Steps implemented by the procedure written in order to analyze the experimental data collected by the bipotentiostat. After data acquisition the procedure computes the Irms value for each measured frequency. Afterwards the program plots the Irms -values as a function of the frequency and fits them linearly. From the fit the value of Cd can be calculated. This sequence can be repeated for every complete data set as, for example, when one wants to measure variations of Cd in time. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 23 2.15 Atomic force microscope: model MFP 3D by Asylum Research. . . . . . . . . . . . . . . . . . . . . . . 24 2.16 Cross section of the mixed SAM formed by ssDNA- and MCH-molecules. MCHmolecules compete with unspecifically bound ssDNA-molecules, removing them and chemisorbing onto the surface through S-Au bond (≈ 2 eV). . . . . . . . . . . . . . . . . . . . . . . . . . . 25 3.1 Root mean squared current, Irms , and its phase, φ, as a function of the applied frequency, f , for a solution of KCl 1 mM and an applied potential, Vrms = 100 mV. The current is shown as red circles, the phase as green squares whereas the blue dashed lines represent the fits obtained using equations 3.1 and 3.2. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 28 3.2 Root mean squared current, Irms , and its phase, φ, as a function of the applied frequency, f , for a solution of KCl 10 mM and an applied potential, Vrms = 100 mV. The current is shown as red circles, the phase as green squares whereas the blue dashed lines represent the fits obtained using equations 3.1 and 3.2. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29 3.3 Impedance, Z, vs frequency curves for KCl at 6 different concentrations, from 1 µM (red dashed curve) to 100 mM (purple dashed line). The salt concentration increases for every profile by one order of magnitude going down from the red curve at the top of the graph. The orange solid line represents the Z-values for a KCl concentration of 10 mM and is the solution of choice for the rest of the experiments in this chapter. . . . . . . . . . . . . . . . . . . . 30 3.4 I vs f profiles as a function of DNA-hybridization (average of five independent measurements, in a 10 mM KCl solution). Red solid line: electrodes functionalized with a SAM of ssDNA + MCH; blue dashed line: same electrodes after hybridization. In the high frequency range the profiles overlap well, indicating that the ionic strength of the solution is the same. Changes in the low frequency range indicate modifications at the electrode-electrolyte interface. Inset: zoom-in of the low frequency range; markers: experimental points, dashed lines: best-linear-fit, same color scheme as main plot. . . . . . . . 31 3.5 Time behavior of Cd for clean, non-functionalized electrodes. Cd seems to reach stability only after a time intervall of 15 h and a percentage change of roughly 25%. . . . . . . . . . . . . . 33 3.6 Relative capacitance change as a function of time for a control measurement (clean electrodes) and two different functionalizations using the two-electrode configuration. Red: bare electrodes, blue: electrodes functionalized with C14 thiols, green: electrodes functionalized with (ssDNA + MCH)-SAM. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 34 100 List of Figures 3.7 Absolute capacitance change as a function of time for three different functionalizations using the two-electrode configuration. Red: electrodes functionalized with a MCH-SAM, green: electrodes functionalized with a (ssDNA + MCH)-SAM, blue: electrodes functionalized with a ssDNA-SAM. On the right of the graph we depicted the situation in three different cartoon. (a) cross section of the SAM formed by the MCH-molecules alone (The alkyl chain is depicted as a zig-zag line, the S-atom and the OH-group by a blue and an orange circle, respectively). (b) SAM composed by the mix ssDNA- + MCH-molecules. (c) SAM composed by the DNA-molecules alone (The ssDNA-molecules are depicted as a sequence of red rods with the thiolated tail (C6)). . . . . 35 3.8 Absolute capacitance change as a function of time measured in KCl 10 mM for the same electrodes functionalized with a C14-SAM after the flux in the channel of different buffer-solutions for 4 h. Black: freshly prepared C14-SAM; blue: after flux of KH2 PO4 ; green: after flux of PBS ; red: after flux of T E at pH = 9. . . . . . . . . . . . . . . . . . . . . . . . . . . . . 37 3.9 Cartoon that emphasizes the irregularities of the SAM (empty areas within the green bars) and clarifies the contribution to Cd given by the multivalent ions (purple and grey circles), that stick on the gold surface after the flux of different buffers in the channel. . . . . 38 3.10 Cartoon of the DDI-process for a SAM of ssDNA (shown on the left) after incubation with the ssDNA-streptavidin conjugates (shown on the right). The ssDNA of the conjugate has the complementary sequence of the immobilized ssDNA on the surface. The conjugates were produced in the laboratories of our collaborator, Dr. Fruk at the Karlsruhe Institut of Technology, Germany. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 39 3.11 Cd vs t profiles as a function of DDI and protein-protein binding in a solution of KCl 10 mM. Blue: electrodes functionalized with a SAM of ssDNA + MCH; red: Cd after hybridization of the DNA-molecules with the complementary ssDNA conjugated with streptavidin-molecules; green: Cd after the interaction of the streptavidin with a solution containing biotinilated aGFAP-molecules, the antibodies for GFAP. . . . . . . . . . . . . 40 3.12 Cd measured in KCl 10 mM as a function of the electrodes functionalization. The values of the differential capacitances were computed averaging the experimental results shown in Figure 3.11 over the last four hours of the experiment. The error is the root mean squared deviation in the same time interval. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 41 3.13 Histograms from [60] where the authors compare the capacitance measured at the interface electrode/electrolyte as a function of the molecular layer on the surface of the electrode. First bar:film of antibodies; second bar:after incubation with albumine; third bar: after incubation with SCCA. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 42 3.14 Cd measured in KCl 10 mM, over a period of 30 h and for a two-electrode-device functionalized with dsDNA-SAM. Time instabilities are clearly visible throughout the measured time-range. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 43 4.1 Scheme of the connections of the three-electrode setup. The potential is applied across WE and RE whereas the current is measured across WE and CE. . . . . . . . . . . . . . . . . . . . . . . 45 List of Figures 4.2 101 Values of the OCP across our Ag/AgCl-RE and a standard RE, Ag/AgCl in saturated KCl solution, for 4 different concentrations of KCl. In the graph we use the nomenclature of Equation 4.1. Red: KCl 100 µM; blue: KCl 1 mM; green: KCl 10 mM; black: KCl 100 mM. The OCP-value was measured for 1 min. Average-values and root mean squared deviations were computed in this time interval and are listed in Table 4.1. . . . . . . . 46 4.3 Root mean squared current, Irms , plotted as a function of the applied frequency, f , for a solution of KCl 10 mM and an applied potential, Vrms = 10 mV. . . . . . . . . . . . . . . . . . . . . . . 47 4.4 Cd vs VBias as a function of DNA-hybridization in a solution of KCl 10 mM. red: electrodes functionalized with a SAM of ssDNA + MCH; blue: Cd after hybridization of the DNA-molecules with the complementary ssDNA. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 50 4.5 contours of the area of the working electrode within which we chose to perform the shaving experiment with AFM. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 50 4.6 Histogram of the heights of the image shown in the inset. The shaving experiment was performed for electrodes functionalized with ssDNA + MCH. Blue line: fit of the histogram performed with gaussian functions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 51 4.7 Histogram of the heights of the image shown in the inset. The shaving experiment was performed for electrodes functionalized with dsDNA + MCH. Blue line: fit of the histogram performed with gaussian functions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 52 4.8 Normalized Cd -change vs time as a function of three different functionalization-layers, in a solution of KCl 10 mM. red: electrodes functionalized with a SAM of MCH; blue: electrodes functionalized with ssDNA + MCH; green: electrodes functionalized with dsDNA + MCH . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 54 4.9 Figure 3.6 with the superimposition of the green plot in Figure 4.8. Red: twoelectrode-setup, bare electrodes; blue: two-electrode-setup, SAM of C14 thiols, green: two-electrode-setup, SAM of ssDNA + MCH; black: three-electrode-setup, SAM of dsDNA + MCH. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 55 4.10 Cd measured in KCl 10 mM using a three-electrode setup with a WE functionalized with dsDNA over a time interval of 11 h. The average value of Cd and its standard deviation over this time interval are: Cd = (0.503 ± 0.004) nF. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 56 4.11 Cd measured in KCl 10 mM vs time as a function of DNA-hybridization. Red circles: WE functionalized with ssDNA + MCH; blue circles: Cd after DNA-hybridization. . . . . . . 56 4.12 Cd measured in KCl 10 mM vs time as a function of SAM formation. Red circles: clean WE; blue circles: WE after the procedure for ssDNA-functionalization. . . . . . . . . . . . . . . . . 57 4.13 Cd measured in KCl 10 mM vs time as a function of DNA-hybridization for a device treated with plasma etching in order to expose the cleanest gold surface possible. Red circles: WE functionalized with ssDNA + MCH; blue circles: Cd after DNA-hybridization. 58 4.14 Cd measured in KCl 100 mM vs time as a function of DNA-hybridization for a device treated with plasma etching in order to expose the cleanest gold surface possible. Red circles: WE functionalized with ssDNA + MCH; blue circles: Cd after DNA-hybridization. 59 102 List of Figures 4.15 Cd measured in KCl 100 mM vs time as a function of DNA-hybridization and with the absence of the MCH-molecules in the SAM. Red circles: WE functionalized with ssDNA; blue circles: Cd after DNA-hybridization. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 59 4.16 Cross section of the assembly composed by gold film, MCH carpet, DNA-molecules and solution. The DNA-molecules are represented as red rods, the MCH molecules are depicted as zig-zag lines on the electrode surface. The sulfur atom is shown as a blue circle while the OH-group as an orange one. Because of the variation in the persistence length of the DNA-molecules after hybridization (from 1 to 50 nm), the ssDNA is composed by 7 small rods whereas the dsDNA by a single straight rod. Also we draw ssDNA with a diameter of roughly half the length of the diameter for dsDNA, as is should be for buffer solutions that completely screen electrostatic charges. The limits of the electrical double layer is shown as a dashed lines. (a) ssDNA-SAM, κ−1 = 1 nm; (b) dsDNA-SAM, κ−1 = 1 nm; (c) ssDNA-SAM, κ−1 = 3 nm; (d) dsDNA-SAM, κ−1 = 3 nm. . . 61 4.17 Calibration curve of the three-electrode device. The response is shown as percentage capacitance variation at the WE as a function of the complementary DNA concentration in solution (KCl 100 mM). The device saturates at a concentration of the target DNA of 100 nM whereas, at 100 pM, the capacitance change is already roughly 9%. . . . . . . . . . . . . 62 4.18 Concentration of the complementary DNA in solution divided by the percentage change of Cd as a function of ccDNA . The graph is plotted in a Log Log scale and the linear fit of the experimental points is shown as a blue dashed curve. From the intercept of the linear function at ccDNA = 0 one can compute the association constant, KA , using Equation 4.4. . . 63 4.19 Study of the kinetics of DNA-hybridization via differential capacitance measurements. The WE of the three-electrode setup was functionalized with a ssDNA SAM and its capacitance vs time was measured in KCl 100 mM. After 30 minutes we exchanged the solution in the pool with one having the same KCl concentration plus 1 nM of the complementary-strand DNA. The green and blue lines represent best-fit models based on first- and second-order Langmuir absorption kinetics, respectively. . . . . . . . . . . . . . . . . . . . . 65 4.20 Cd measured in KCl 100 mM vs time as a function of DNA-hybridization and with the presence of the TOEG6-molecules in the SAM. Red circles: WE functionalized with ssDNA + TOEG6; blue circles: Cd after DNA-hybridization. . . . . . . . . . . . . . . . . . . . . . . . . . 67 5.1 Pictorial view of the three main parts of the device for parallel detection of different sequences of DNA within a microchannel. a) base plate constituted by a glass slide with the three couples of WE and CE. b) PDMS membrane with microchannel. c) Ag/Ag/Cl lamina which serves as RE and upper wall of the channel. Holes are drilled in the lamina in order to connect the microchannel with the pump-system. . . . . . . . . . . . . . . . . . . . . . . . . . 73 A.1 Model of the electrode/electrolyte interface. The ions in solutions can either stick on the surface, within the inner Helmholtz plane or remain in the so called diffusive layer, beyond the outer Helmholtz plane. In the diffusive layer the ions reorganize according to Boltzmann’s distribution. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 78 List of Figures 103 A.2 Potential profile, φ, as a function of the distance from the electrode, x. The distance x2 corresponds to the IHP. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 79 A.3 Theoretical behavior of Cd as a function of the applied voltage E at the electrode. For very positive and negative voltages the Stern’s layer dominate and the capacitance is constant. At potentials equal to the point of zero charge, the potential for which the attached ions at the surface counterbalance the charge density of the electrode, the capacitance has a parabolic shape generated by the diffused ions . . . . . . . . . . . . . . . . . . . . . . 79 B.1 Picture of the spin-coater used during the thesis-work. Its main features are listed in Table B.1. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 81 B.2 Picture of the mask aligner MJB3 used during the thesis-work. Its main features are listed in Table B.2. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83 B.3 Picture of the mask aligner MA25. Its main features are listed in Table B.2. . . . . . . . . . . . . . 83 B.4 Picture of the e-beam evaporator used during the Ph.D. Its main features are listed in Table B.3. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 85