WO2016054222 - Auburn University
advertisement

(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT)
(19) World Intellectual Property
Organization
International Bureau
(43) International Publication Date
7 April 2016 (07.04.2016)
§IJ
~
WIPO I PCT
(51) International Patent Classification:
AOIN 63100 (2006.01)
A61K 35174 (2015.01)
(21) International Application Number:
PCT/US2015/053239
(22) International Filing Date:
30 September 2015 (30.09.2015)
(25) Filing Language:
English
Illlll llllllll II llllll lllll lllll lllll llll III Ill lllll lllll lllll 111111111111111111111111111111111
(10) International Publication Number
WO 2016/054222 Al
AO,AT,AU,AZ,BA,BB,BG,BH,BN,BR,BW,BY,
BZ,CA,CH,CL,CN,CO,CR,CU,CZ,DE,DK,DM,
DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT,
HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR,
KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG,
MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM,
PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC,
SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN,
TR, TT, TZ, VA, VG, US, UZ, VC, VN, ZA, ZM, ZW.
(26) Publication Language:
(30)
(71)
(72)
(71)
English (84) Designated States (unless otherwise indicated, for every
kind of regional protection available): ARIPO (BW, GH,
Priority Data:
GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ,
62/057,667 30 September 2014 (30.09.2014)
us
TZ, VG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU,
Applicant: AUBURN UNIVERSITY [US/US]; 570
TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE,
Devall Drive, Auburn, AL 36832 (US).
DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU,
LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK,
Inventors; and
SM, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ,
Applicants : LILES, Mark, R. [US/US]; 710 Sanders
GW, KM, ML, MR, NE, SN, TD, TG).
Street, Auburn, AL 36830 (US). KLOEPPER, Joseph
[US/US]; 735 S. Gay St., Auburn, AL 36830 (US).
Published:
(74) Agents: MCBRIDE, Scott, M. et al.; Andrus Intellectual
Property Law, LLP, IOO East Wisconsin Avenue, Suite
l IOO, Milwaukee, WI 53202 (US).
=
=
-------
with international search report (Art. 21(3))
with sequence listing part of description (Rule 5.2(a))
(81) Designated States (unless othenvise indicated, for every
kind of national protection available): AE, AG, AL, AM,
;;;;;;;;;;;;;;;
;;;;;;;;;;;;;;;
;;;;;;;;;;;;;;;
;;;;;;;;;;;;;;;
;;;;;;;;;;;;;;;
;;;;;;;;;;;;;;;
(54) Title: USE OF PECTIN OR PECTIN-RELATED SACCHARIDES TO ENHANCE EFFICACY OF PLANT GROWTH-PROMOTING RHIZOBACTERIA (PGPR) STRAINS FOR PROMOTING GROWTH AND HEALTH IN PLANTS AND ANIMALS
(57) Abstract: Disclosed are compositions and methods that include or utilize plant growth promoting xhizobacteria (PGPR) for improving growth and health in plants and animals. The compositions and methods include or utilize a plant growth promoting
rfiizobacteria (PGPR) that expresses a protein associated with pectin metabolism, and a saccharide comprising pectin or a pectin -re lated saccharide.
WO 2016/054222
PCT/US2015/053239
USE O.F PECTIN OR PECTIN-RELATED SACCHi-\RIDES TO
.ENHANCE EFFICACY OF PLANT GRO\VTH-PROJ\'10TlNG
RHIZOBACTERIA (PGPR) STRAINS FOR PROlVIOTlNG GRO\VTH
AND HKALTH IN PLANTS AND ANll\lALS
CROSS-REFERENCE TO RELATED A,PPLICAT£0NS
[0001]
The present application dairns the benefit of priorit,y under 35 lJ.S .C § 119(e)
to U.S. provisional application No. 62/057,.667, filed on September 30, 2015,. the content of
which is incorporated herein by reference in its ~mt.irety.
FJELD
[0002]
The presently disdosed subject 1natter relates to the field of plant grov.·th-
promoting rhizohacteria (PGPR). In particular, the present subject t.natter relates to the use of
pectin or pectin-related saccharide to enhance the efficacy of PGPR in regard to promoting
growth and heahh in plants and aninrnls_
BACKGROUND
[0003]
Pl.ant-associated microorganisms have been extensively
exarnim.~d
for their
roles in natural and induced suppressiveness of soilborne diseases. Among the many groups
of such organisms are root-associated bacteria, ·which generally represent a subset of soil
bacteria_ Rhizobacteria are a subset of total rhizosphere bacteria \.vhich have the capacity,
upon re-introduction to seeds or vegetative plant parts (such as potato seed pieces), to
colonize the developing mot systen1 in the presence of cmnpeting soil rnicroflorn_ Root
colonization is typically examined by quantifying bacteiial populations on root surfaces;
hcnvever, some rhizobacteria rnn also enter roots and establish at least a !irnited endophytic
phase. Hence, root colonization may he viewed as a continuum frorn the rhizosphere to the
rhizoplane to internal tissues of roots.
[0004]
Rhizoba<.:teria \Vhich exert a beneficial effect on the plant being coloniwd a.re
tenned "plant-grrn.vth promoting rhizohacteria" or "PGPR." PGPR may benefit the host by
1
WO 2016/054222
PCT/US2015/053239
causing plant gro\vth promotion or biological disease controL The sa1r1e strnin of PGPR may
cause both growth promotion and biological control.. Among the soilborne pathogens sho\vn
to be negatively affected by PGPR are Aphanomyces spp., Fusarium m:mporum,
Gaeumannon1yct.w graminis, PhJ'tophthon.1 spp., f>ythium spp., Rhizoctonia solani, Sclero!ium
ro(t:~·ii,
lhielaviopsis basicola, and Verticillium spp. In most of these cases,, biological control
results frmn bacterial production of metabolites \vh1ch directly inhibit the pathogen, such as
antibiotics, hydrogen cyanide, iron-chelating siderophores, and cell ·wall-degrading enzymes.
Plant growth promotion by PGPR nrny also be an indirect mechanism of biological control,
leading to a reduction in the probah1!ity of a plant contracting a disease 'Nhen the grmvth
promotion results in shortening the time that a plant is in a susceptible state, e.g. in the case
,.vhere PGPR cause enhanced seedling emergence rate, thereby reducing the susceptible time
fbr pre-ernergence damping-off An alternative n1echanisrn for biological control by PGPR is
induced systemic resistance. PGPR and uses thereof are disclosed in the prior art. (See. e.g.,
tLS. Patent Nos. 8,445,255; 6,524,998; 5,935,839; 5,640,803; 5,503,652; and 5,503,651; the
contents of which are incorporated herein by reference in their entirety).
[0005]
In addition to their observed association in naiure \Vith plants, PGPR also rnay
be utilized as probiotics for animals in order to improve animal gro\vth or animal health. For
example, Bacillus amyloliqinf(u.·iens subsp. plamarwn AP 193 has been described as a
prnbiotic for fish. (See U.S. Published 1\pplication No. 2012/03285 72).
[0006]
In swine, probiotics have been used to have a positive influence on gut
microbiota balance, intestinal epithelium integrity and maturation of gut-associated tissue.
(See Corcionivoshi
el
al., Animal Science and Biotechnologies, 20 l 0, 43(1 ))_
In poultry,
prnbiotics have been used to maintain digestive microbial balance and to reduce potential
pathogenic bacteria ·1Nhich results in improving grmvth., egg production, and teed conversion_
(See id).
In cattle, prohiotics have been used to prevent and combat digestive disorders such
as diarrhea during lactation, to influence ruminal metabollsm of nutrients, \vhich helps
maintain health and improve productive performance. (See id). In sheep, probiotics have
been used to prevent and combat pathological conditions that aiise from digestive balance.
(See id).
2
WO 2016/054222
[0007]
PCT/US2015/053239
Therefore, ne\v compositions a11d methods of use for PGPR m promoting
grmvth and health 1n plants and animals are desirable.
SUlVlM,i\RY
[0008]
Disclosed are compositions and rnethods that include or utilize plant grnwth
promoting rhizobacteria (PGPR) for improving gro\vth and health in plants and animals. The
cornpositions and methods include or utilize a plant growth promoting rhizobacteria (PGPR)
that expresses a protein associated 'vith pectin metabolism, and a saccharide comprising
pectin or a pectin-rdawd saccharide.
[0009]
The disclosed compositions may include inoculants which comprise: (a) a
plant gnJ\.Vth promoting rhizobacteria (PGPR) that expresses a protein associated \Vith pectin
metabolism; and (b) a saccharide comprising pectin or a pectin-related saccharide. Suitable
PGPR may include Bacillus species such as Bacillus amylo!iqu<c:fhciens subspecies
planlanun. The pectin or pectin-related saccharides may include pectin-derived saccharides
such as hydrolyzed pectin, D-galacturonate, D-glucuronaie, or mixtures thereof Optionally,
the pect1n or pectin-related saccharide fi.1nctfrms as a carrier for the PGPR and.ior the inocu!ant
includes a carrier other than the pectin or pectin-related saccharide.
[0010]
The disclosed compositions may· be used to treat plants, seeds, and soils in
order to i1nprove plant growth or plant health. The disclosed compositions may be fommlated
as a plant treatment composition, a coating for seeds, or a soil amendment composition.
[0011]
The disclosed compositions also may be administered to animals in order to
improve animal gr<.)\.Vth or anirnal health. The disclosed cornpositions ma)/ be .formulated as
an animal feed, such as a pelleted animal feed.
[0012]
Also disclosed are inetlmds of using pectin or pectin-related saccharides to
improve the efficacy of PGPR in regard to promoting i:,•rowth or health in plants and animals.
The disclosed methods for i1nproving plant growth or plant health may include: (a) treating
plants, seeds, or soi! \.Vith a plant grmvth promoting rhizobacteria (PGPR) that expresses a
3
WO 2016/054222
PCT/US2015/053239
protein associated with pectin metabolism and (b) treat1ng the plants, seeds, or soi! "\Vith a
saccharide comprising pectin or a pectin-related saccharide, \vhere the plants, seeds,, or soil
may be treated with the PGPR ai1d the sacchadde co11currently or are treated \vith the PGPR
and saccharide non-currently in either order. The disclosed methods for improving animal
gro\:vth or animal health may include (a) administering to an animal a plant grO\vth promoting
rhizobacteria (PGPR) that expresses a protein associated with pectin metabolism and (b)
administering to the a11imal a sacchari<le comprising pectin or a pectin-related saccharide,.
vd1ere the animals may be administered the PGPR and the saccharide concurrentl.y or are
treated \Vith the PGPR and saccharide non-currently in either ordeL
[0013]
Also disclosed are methods of using pectin or pectin-related saccharides to
prepare compositions and inoculants as disclosed herein.
The methods may include
cornbining PG PR and pectin, vd1ich has been extracted from pectin-containing plant 1naterial,
or pectin-related saccharides to prepare the disclosed compositions and inoculants.
Optionally, a carrier may be combined with the PGPR and pectin or pectiiH·elated
saccharides to prepare the disclosed compositions and inoculants.
BRIEF DESCRIPTION OF THE FIGURES
[0014]
Figure I. Phylogeny of PGPR Bacillus spp. evaluated in Exmnple 1. (Panel A)
Neighbor joining phylogenetic tree based on gyrR sequem:es using B. cereus ATCC l4579T
as an outgroup. (Panel B) J'vlaximum-likelihood phylogenetic tree of the 25 B. subtilis group
strains based on 729,383 bp sequence of core genome. Tw·o dusters belonging to H
amyloliquefaciens snhsp. plantarurn and H. wnyloliquejaciens subsp. amy!oliquefaciens are
indicated by brackets.
[0015]
core
gt~nomes
Figure 2. The distribution of difforent subsystem categories of four different
specific to genus Bacillus (n=8 ! ), B. suhtilis subgroup (n=53), species H
amylo!iquefaciens (n=32) m1d subsp. plantarum (n""28). (Panel A) The total counts for genes
within difforent subsystem categor1es for each of the core genomes. (Panel B) The 0..{i relative
abundance of the genes within different subsystem categories tor each of the core genomes.
4
WO 2016/054222
PCT/US2015/053239
(Panel C) Categories of functions encoded by the 73 B. amyloliqutf'aciens subsp. plamarwn-
spedfic genes present il1 the B. am.yloliquefaciens suhsp. plantarum core genome but absent
in the B. amy!oliqucdcK'iens species-level core genome. The number beside each subgroup of
the pie figure represents fae number of genes encoding the function.
[0016]
Figure 3. Antilnicrobja] actjvities of Bacillus sp. AP l 93 and .its mutants
l\,'i1_f4A, defoctive in surfactin expression, t\c{fi1D, defocfrve in difficidin expression, and A4j1_.
defective hi the expression of multiple secondary metabolites (including diffic1din) against
plant pathogens Pseudornonas
.~yriuge
pv. tabaci, Rhizobium radiobacter, .){anthomonas
axonopodis pv. vesicatoria and )(anthomonas axonopodis pv. l'.ampestris as demonstrated
\.Vith an agar diffusion assay.
[0017]
Figure 4. LC-i'v1S spectra for metabolites from cell-free supernatants of (A)
wild-type B. amyloliqm..;li:wiens AP 193,. and (B) its isogenic <:f.fnD mutant, \·vhen grovm in TSB
for 72 hours. Note that in negative ion niode that only the dcprotonated fonn of oxydifficidin
\.Vas detected in bacteria! culture supernatants at a m./z 5593.
[0018]
Figure 5. Expression of a pectin l.yase m:.:tivity by PGPR Bap strain APl93.
Now the ckared halo around the growth of the Hap strnin due to pectin degradation.
[0019]
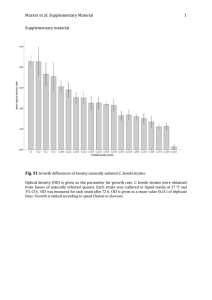
Figure 6. Use of l ci/o pectin as a sole C source by PGPR st.rains AP143 and
AP 193 in TSS medium. The small increase in
ODr;i~;
by the non-PGPR strain HD73 vrns due
to residual nutrients present from the previous TSB culture.
DETA lLED DESCRIPTION
[0020]
The disclosed subject matter of the invention may be described using various
terms as described belo-.;.v.
[0021]
Unless othenvise spcdfied or indkatcd by context, the tcnns "a", "an'·', and
"the" mean "one or more." For example, "a sugar" should be interpreted to mean "one or
more sugars" unless othenvise specified or indicated by wntext
5
WO 2016/054222
[0022]
PCT/US2015/053239
As used herein, "about", "approximately," "substantially,'' and "significantly"
\'-'ill he understood by persons of ordinary skill in the art and •vill vary to some extent on the
context in \vhich the),, are used. ff there are uses of the tenn which are not dear to persons of
ordinary skill in the art given the context in w"hich it is used, ''about" and ''approxhnately"
\:vil! mean plus or minus :S l OO.··r1 of the particular tenn and "substantially" and "significantly''
·\viU mean pl us or minus > 101hi of the particular term.
[0023]
As used herein, "about", "approximately,'' "substantially," and "significantly''
'vii! be understood by persons of ordinary skill in the art and will vary to some extent on the
context in \Vhich they are used. If there are uses of the term \vhkh are not dear to persons of
ordinary skill in the att given the context in which it 1s used, "about" and "approximately"
will mean plus or minus 5:10°,...ii of the particular term and "substantially" and "sit,>nifica11tly"
\-viii rnean plus or minus > 100.:{, of the particular term.
[0024]
As used herein, the tenns "include" and "including" have the s;m1e meaning as
the tem1s "comprise" and "comprising." The tenns "comprise" and "comprising" should be
interpreted as
bt~ing
"open" transitional tem1s that permit the inclusion of additional
components further to those components recited in the claims.
The terms "consist" and
'"consisting of' should be interpreted as being "closed" transitional terms that do not permit
the inclusion of additional components other than the components recited in the claims. The
tem1 "consisting essentially of' should be interpreted
to
be partially closed and allowing the
inclusion only of additional cornponents that do not fundamentally alter the nature of the
claimed subject matter.
[0025]
The term "plant" as utilized herein should be interpreted broadly and may
include angiosperms and gymnospem1s, d:icots and monocots, and trees.
Examples of
angiospem1 dkots may include, but are not limited to tomato, tobacco, cotton, rapeseed, field
beans, soybeans, peppers, lettuce, peas, alfa.lfo,
dov,~r,
cabbage, broccoli, cauliflower, brussel
sprouts), radish, carrot, beets,. eggplant, spinach, cucumber, squash, melons, cantaloupe, and
sunflowers, Example of a:ngiospenn monocots may include, but are not limited to asparagus,
field and sweet com, barley, ·wheat, rice, sorghum, onion, pearl millet. rye, oats, and sugar
6
WO 2016/054222
PCT/US2015/053239
cane. \:Voody plants nuiy include, but are not limited to fruit trees, acada, alder, aspen, beech,
birch, sweet gum, sycamore, poplar, willow, fir, pine, spruce, larch, cedar, and hemlock.
[0026]
The term "anirnar' as utilized herein should he interpreted broadly and may
include mammals and non-mammals.
f'vlamrnals may include human and non-human
mannnals, such as cows, pigs, sheep, and the like. Non-mamma!.s may include birds (e.g.,
chickens, turkeys, ducks, and the like) and fish.
[0027]
The present inventors have identified a collection of plant grm.vth-promoting
rhizobacteria (PGPR) that are capable of improving the growth of plants, and also have
disease- and pest-controlling activ1ty. From an analysis of genome sequences from the hestperforming Bacillus amyloliqut;f(-1ciens subspecies plamarum PGPR strains, the inventors
identified some genetically encoded fi.1ncth-ms that are always present \Vlthin these Bacillus
PGPR strains and are not present in other Bacillus species that are not plant-related. In
p<trticular, these PGPR strains can use sugars derived from plant pectin as a carbon m1d/or
energy source. By supplementing pectin on plant seeds that are inoculated with Bacillus
spores, or by supplementing the an1ount of pectin available for Bacillus PGPR strain postseed germination, this \viii result in an enhancement of l) the Bacillus strain colonization of
the plant rhizosphere and/or 2) better p('rsistence of Bacillus v.dthin the plant rhizosphere
and/or 3) better plant grO\vth performance in response to PGPR strain
+ pectin adm.inistration
amlior 4) better biological contrnl of disease (e.g., bacteria, fungi, viruses) or pests (e.g.,
nematodes) as a result of PGPR strain + pectin adminisirntion.
[0028]
[0029]
The term "plant gnnvth promoting rhizobacteria" or "PGPR" reJers to a group
of bacteria that colonize plant roots, and in doing so, promote plant growth and/or reduce
disease or damage from predators. Bacteria that are PGPR rnay bdong to genera including,
hut not
limited to Actinobacter,
A!ca!igenes, Bacillus..
Burkholdcria,
Buttiauxel!a,
Enterobacter, K!ebsiella, Kluyvera, Pseudomonas, Rahne/la, Ralstonia, Rhl.zobiwn, Serraria,
Swnotrophomonas. Paenibacil!us, and Lysinibacillus. The PGPR utllized in the disclosed
7
WO 2016/054222
PCT/US2015/053239
methods and composition nrny be a single strain, speCH.$, or genus of bacteria or rnay
comprise a mixture of bacterial strains, species, or genera. For example, the PGPR may be
selected from genern including, but not limited to, Aclinobacter, Alcaligenes, Bacillus.
Burkholderia, Buttiauxefla, Enterobacter, Klebsiel!a, Kluyvera, Pseudomonas, Ralmel!a,
Ralsionia. Rhizobium, Serratia, Stenotrophomonas, Paenibacil/us, and LJwinibacillus.
[0030]
The genus Bacillus as used herein refors to a genus of
Gram-positiv~\
rod-
shaped bacteria which are members of the division Firmicutes. Under stressful environmental
conditions, the Bacillus bacteria produce oval endospores that can stay dormant for extended
periods.
Bacillus bacteria may be characterized and identified based on the nudeotide
sequence of their 16S rRNA or a fragment thereof ( e.g, approximately a 1000 nt, 1100 nt,
1200 nt, 1300 nt, 1400 nt, or 1500 nt fragment of l 6S rRNA or rDNA nucleotide sequence).
Bacillus bacteria xnay include" bui are not fonited to B. acidiceler, B. acidicola, B.
acidiproducens, B. aeoli11s, B. uerius, B. uerophilus, B. agaradhaerens.. B. uidingensis, B.
akibai, B. aicalophilus. B. algicola, B. alkalinitrilicus, B. alkalisediminis, B. alkalitelluris, B.
altitudinis, B. alvazyuensis. B. amyloliq1wfi:1ciens. R anthracis, R aquinwris, B. arsenicus, B.
1.uyahhattai, H asahii, B. atrophaeus, H. aurantiacus, B. azoNd(mnans, B. hadius, H.
harbaricus, 13. hataviensis. B. bet;ingensis, B. benzoevorans, H bevericz<z,ei, B.
bogoriensl~,
B.
bomniphi/us,. B. bwanolivorans, R canaveralius, B. carboniphihts, B. cecembensis, B.
cellulosizvticus. B. l·ereus. B. c:hagannorensis, B. chungangensis, H cib/. B. circulans. H
c!arkii. B. dausii, B. coagulans, B. coahuiiensis, B. colmii, B. decis{fhmdis. H. deco/orationis,
B. drentensis, B. farraginis. B. fastidiosus . B. jirmus, B. ./lexus, B. fhraminis . B. j(wdii, B.
fiN·tis, B. jiunarioli. B. jimicu/us, B. galactosidizyticus, B. galliciensis, B. ge/atini.. R gibsonii,
H. ginsengi, B. ginsengihumi, B. grarninis, B. halmapalus, H. halochares, H. halodurans, H.
hernicellulosi(vticus. B. herbertsteimmsis, B. horikoshi, B. homeckiae, B. horti, B. humi, IJ.
hwqjinpoensis, B. idriensis, B. indicus, H if?famis. B. it!lemus, B. isaheliae, B. isronensis, B.
jeotgali. B. koreensis, B. korlensis. B. krihbensis, B. krulwichiae, B. lehensis, B. lenius. B.
licho1{fhrmis, R lilora/is, B. locisalis, B. luc{/('rensis, B. luteoius., B. macauensis, B. ma(.}'ae,
B. mannanizvticus. B.
nwrL~flavi,
B. marrnarensis, 8, nw.Ysiliensis, B. megaterium, H.
methanolicus, B. meihylotrophicus, B.
tn<~javensis,
8
H. rnuralis, B. murifnartini, B. tnJ'Coides, H.
WO 2016/054222
PCT/US2015/053239
nanhaiensis, B. ncmhaiisediminis, B. neat:'>onii, B. neizhouensis, B. niabensis, B. niacini, B.
nova/is, B. oceanisedirninis. B.
oshimensis,
Ol~lwse_yi,
B. okhensis, B. okuhidensis. H oleronius. H
B. panacilerrae, B. paragoniensis, B. persepo!ensis, B. p/akorti<.:fo',
pocheonensis, B.
po~ygoni.
B. pseudoalcaliphilus, B.
pseud(~finnus,
B.
B. pseudomycoides, B.
psychrosaccharof_viicus, B. pumilus, B. qingdaonensis, B. rigui, B. ruris, B. .wknsis.. B.
sa!arius,
B.
saliphi!us,
B.
schlege!ii,
B.
selenafw:~·enatis,
B.
selenitireducens,
B.
seohaeanensis, B. shack!etonii, B. siamensis, B. simplex, B. siralis. B. smithii, B. soli, 8.
solisalsi, B. sonorensis, B. sporothermodurans, B.
sfrato,~phericus,
B. subterraneus, B.
subtilis, B. taeansis. B. tequilensis, B. 1hermantarcticus, B. thennoarn.vlovorans, B.
thermocloacae, B. fhermolactis, B. thioparans, B. thuringiensis, B. fripox.ylicola, B. tusciae,
B. vallismorlis, B. vedderi. B. vietuamensis. B. vireti, B. >vakoensis. B. weihensiephanensis.
B. xiaoxiensis, and mixtures or blends thereof
[0031]
The PGPR and inoculants thereof disclosed herein may include H
am_y!oliq1u.'.fi.r.ciens or a Bacillus species that is closely related to /{ amyloliquefaciens. A
Bacillus species that is dosel:y· related to B. amyloliqm:'.fhcieus may be defined as a species
having a 16S rDNA sequence comprising SEQ ID N0:26 or comprising a 16S rDNA
sequence having at least about
98~/!}
or 99%.i sequence identity' to SEQ ID N0:26. The PGPR
preferably is B. arnyloliq1u.faciens subspecies plamarwn or a Bacillus species that 1s closely
related to B. mnyloliqur;:fhciens subspecies p!antarum. H amylo!iqu,fl-Jciens subspecies
plaiHannn 1s a subspecies of B. amy!oliqu4hciens which is colonizes plant roots and typically
exhibits axny'lase activh:y, Suitable PGPR stra,ins for the disclosed rnethods and compositions
may include PGPR strains having a gyrB gene that exhibits sequence identity to the gyrB gene
from strains of Bacillus mnyloliquefaciens subspecies p!antarum. In some embodiment, the
PGPR strain utilized in the disclosed methods and co1npositions has at gvrB gene having at
least about
80~'o,
90%, 95 1J...ii, 96t:.o,
97~{1,
98 1l.·ii, 99% or I OQtl,/(, sequence identit)" to the
polynudeotide sequence of SEQ ID N0:25,. \vhich is the polynucleotide sequence of the gyrB
gene from strains of Bacillus amyloliqmfaciens subspecies plan/arum.
[0032]
Suit.able st.rains of B. amyloliqw(/{:.1ciens subspecies plantarwn for use in the
disclosed cmnposiHons and xnethods rnay include but are not lilnhed to Bacillus
9
WO 2016/054222
PCT/US2015/053239
a.myloliqu4hciens snbsp . p!antarum AS43.3, Bacillus amy!o!iquejaciens snbsp, plantarum
TrigoCorl 448,
Bacillus amyloliquefac:iens
amyloliquef(:1ciens subsp. pkmtarum
plantarwn
EBL l l,
Bacillus
suhsp,
UCMB5 ! 13,
arnyloliquejaciens
p/antarum
UC:rv1B5033,
Baci//u,<;.·
Bacillus amyloliqutfaciens subsp.
subsp.
plantarum
\\/2,
Bacillus
tnnyloliquefdciens subsp. plammwn lJCf\·1B5036, Bacillus runyloliquefaciens subsp.
planlarum IT-45, Bacillus amy!oliq1ufi.lciens subsp. plantarwn UASWS BA 1, Bacillus
atnJ1oliquef(u.·iens subsp. pfantarum LFB ! l 2, Bacillus amyloliqm.faciens subsp, plamarum
CAUB946, Bacillus
amy!oliquf:.fi.~ciens
subsp. plantarum lVl27, Bacillus amyloliqut.fl:tciens
subsp. plan ta rum B 1895, Bac:i/lus amyloliquefaciens subsp. p/cmtmwn SQR9, Bacillus
an1yloliqw.:f'acie11s subsp_ planiarum AH! 59-l, Bacillus amyloliqut;fcJciens subsp. plan/arum
DC-l2,
Bacillus
amvlo!iauef(.1cien~·
..·
}
~
subsp.
p!cmtarum
YAU
B9601-Y2,.
Bacillus
amyloliq1.1r:.:fc1cie11s subsp. plamamm Y2, Bacillus amyloliquefl".tciens subsp. plamarum
EGD ___AQ14.
Bacillus
amyloliquefaciens
subsp.
plantarum
NAU-B3,
Bacillus
amyloliqmfaciens subsp. plantarwn FZB42, Bacillus amyloliqtt(faciens subsp. phmtarum
CCl 78, Bacillus amylolique,/aciens subsp. plantarum AP79, Bacillus amyloliquefaciens
subsp. plantarum AP7 I, Bacillus amyloliquefi.Jciens subsp. plan ta rum AP 143, Bacillus
mnyloliq1R:fc1cie11s subsp. plan/arum AP193, Bacillus amy!oliq1ufilciens subsp. plantarum
ABOl, and Bacillus amyloliqur:.;{i.lciens suhsp. plantan1m GB03.
[0033]
Suhable PGPR strains and inoculants
thereof for the methods and
compositions disclosed herein may includ(' PGPR strains that express one or rnore proteins
associated with pectin rnetabolisn1. ln some ernbodhnents, the PGPR strnin may express one
or more proteins associated with pectin metabolism, \Vhich may include but are not limited to
proteins encoded by a gene selected from the group consisting of uxaA (a!tronate
dehydratase), uxaB (a!tronate oxidoreductase), uxaC (uronate isomerase), uxaA fmannonate
dehydratase, uxuB (D-mannonate oxidoredm.:tase), kdgA (4-hydroxy-2-oxoglutarate a!do!a.se),
kdgK (2-dehydro-3-deoxyg!uconokinase), exuR (hexuronate utilization opemn transcriptional
repressor), exuT (hexuronate trnnsporter), and cornbinatfons thereof In some embodiments,
the PGPR strain may express one or more pectinase enzymes selected fron1 a group consisting
of pectin lyase, pectate lyase, polygalacturonase, and pectin esterase,
10
WO 2016/054222
[0034]
PCT/US2015/053239
The uxaA gene encodes an ~mzyme whk:h is an altronate dehydratase
(EC:4.2. L 7) \vhich converts D-a!tronate to 2-dehydro-3-deoxy-D-gluconate and \vater.
Therefore, suitable PGPR strains and inoculants thereof for the rnethods and composition
disclosed herein may include a PGPR strain that expresses altronate dehydratase. SEQ ID
NO: l provides the polynudeotide sequence encoding fbr altronate dehydratase.
SEQ ID
N0:2 provides the amino acid sequence frx altronate dehydrntase.
[0035]
The uxaB gene encodes an enzy1ne which is an aim.mate oxidoreductase
(EC:5.3J.12) which converts D-altronate and NAD; to D-tagaturonate and NADR
Therefore, suitable PGPR strains and inoculant:s thereof k)r the rnethods and cornposition
disclosed herein may include a PGPR strain t11at expresses altronate oxidoreductase. SEQ ID
N0:3 providt$ the polynudeotide sequence encoding for altronate oxidoreductase. SEQ ID
NOA provides the an1ino acid sequence for alironate oxidorednctase.
[0036]
The uxaC' gene encodes an enzyme \Vhich is an uronate 1somerase
(EC:! 3.1.12) \vhich converts D-glucuronate to D-fructuronate and w"hich converts D-
galactnronate to D-tagat.uronate. Therefore, suitable PGPR strains and inoculants thereof for
the methods and composition d1sdosed herein may include a PGPR strain that expresses
uronaw isomernse..
SEQ lD N0:5 provides the polynndeot1de sequence encoding for
altrnnate oxidoreductase.
SEQ ID N0:6 provides the amino acid sequence for altronate
oxidoreductase.
[0037]
The uxuA gene encodes a11 enzy.111e "vhich is a mannonate dehydratase
(EC:4.2. L8) vd1ich converts D-rnannonate to 2¥dehydro-3-deoxy¥D-gluconate.
Therefore,
suitable PGPR strains and inoculants thereof for the r:net:hods and composition disclosed
herein may indude a PCJPR strain that expresses mannonate dehyd.ratase.
SEQ ID N0:7
provides the polynudeotide sequence encoding for mannonate dehydratase. SEQ ID N0:8
provides the amino acid sequence for mannonate ckhydratase.
[0038]
The uxuB gene encodes an enzyme \Vhich is a D-rnannonate oxidoreductase
(EC:LLl.57) which converts D-rmmnonate and Nl\.D
11
to D¥fructnronate and NADH.
WO 2016/054222
PCT/US2015/053239
Therefbre, suitable PGPR strnins and inoculants thereof for the
m(~thods
and composition
disclosed herein may include a PGPR strain that expresses D-mannonate oxidoreductase,
SEQ ID N0:9 provides the polynm::Ieotide sequence encoding for altronate oxidoreductase.
SEQ ID NO: 10 provides the amino acid sequence for altronate oxidoreductase.
[0039]
The kdgA gene encodes an enzyrne which is a 4-hydroxy-2-oxoglutarate
aldolase (EC 4. L3, l 6) \.Vhich converts 4-hydroxy-2-oxoglutarate to pyruvate and glyoxylatc,
and \vhich converts 2-dehydro-3-deoxy-6-phosphate-D-gluconate to pyrnvate and Dglyceraldehyde 3-phosphate. Therefore, suitable PGPR strains and inoculants thereof for the
methods and composition disclosed herein may
hydroxy-2-oxoglutarate aldolase,
includ(~
a PGPR strain that expresses 4-
SEQ ID NO: 11 provides the polynucleotide sequence
encoding for 4-hydroxy-2-oxoglmarate aldolase.
SEQ ID NO: 12 provides the amino acid
sequence for 4-hydro.xy-2-oxoglutarate aldolase.
[0040]
The kdgK gene encodes an enzyme \>Vhich is 2-dehydro-3-deoxyg!uconokinase
(EC 2. 7.1.45) \vhich phosphorylates 2-keto-3-deoxygl.ut:onate (KDG) to produce 2-keto-3deoxy-6-phosphogluconate (KDPG). Therefore, suiiab!.e PGPR strains and inoculants thereof
for the methods and composition disclosed herein may include a PGPR strain that expresses
2-(khydro-3-deoxygluconokinase.
SEQ ID NO: 13 provides the po!ynudeotide sequence
encoding for 2-dehydro-3-deoxygluconokina.se.
SEQ ID NO: 14 provides the amino acid
sequence for 2-dehydro-3-deoxyglnconokinase.
[0041]
repressor.
The cafR gene encodes a hexuronatc utilization operon transcriptional
Therefore, suitable PGPR strains and inoculants thereof for the methods m1d
composition disclosed herein may include a PGPR strain that expresses a hexuronate
utilization operon transcriptional repressor.
SEQ ID NO:l5 provides the polynncleotide
sequence encoding for a hexuronate utilization operon transcriptional repressoL
SEQ ID
NO:l6 provides the arnino acid sequence for a hexuronate utilization operon transcriptional
repressor.
12
WO 2016/054222
[0042]
PCT/US2015/053239
The exuT gent~ encodes a hexuronate transporter whkh exhibits hexuronate
transmembrane transporter activity. Therdbre, suitable PGPR strains and inoculants thereof
for the methods and composition disdosed hereiu may iudude a PGPR straiu that expresses a
hexuronate transporteL SEQ ID NO: 17 provides the polynudeotide sequence encoding for a
hexuronate transporter_ SEQ ID NO: 18 provides the amino acid sequence for a hexuronate
transporter.
[0043]
In son1e en1bodiments, the PGPR strain may express one or more pectinase
enzymes selected from a group consisting of pectin lyase (EC 4.2.2. l 0), pectate lyase (EC
4.2.2.2), polygalacturonase (EC 3.2.L"l5), and pectin est.erase (EC 3.1.1.ll). SEQ lD NO:l9
provides the polynucleotide sequence encoding for a pectate !yase precursor. SEQ ID N0:20
providt.$ the amino acid sequence for a pt.•ctate lyase precursor, SEQ ID N0:21 provides the
polynudeotide sequence encoding for a pectin-lyase like protein. SEQ ID N0:22 provides
the amino acid sequence for a pectin-lyase like protein.
SEQ ID N0:23 provides the
polynudeotide sequence encoding for a pectin lyase. SEQ ID N0:24 provides the an1ino acid
sequence for a pectin lyase.
[0044]
of equivalent
"Percentage sequence identity" may be determined by aligning t<.vo sequences
k~ngt.h
using the Basic Local Ahgmnent Search Too! (BLAST) available at the
National Center for 81otechnology lnformation (NCBl) website (i,e., "'bl2seq" as described in
Tatiana A. Tatnsova, Thomas L !'vladden
0 999),
"Blast 2 sequences - a new tool for
comparmg protein and nucleobde sequences", FE!vlS !v1icrobiol
Lett. 174:247-250,
incorporated herein by reference in its entirety), For example, percentage sequence identity
between SEQ ID NO: 1 and another sequence for cmnparison tnay be detennined by aligning
these tviiO sequences using the online BLAST software provided at the NCBI \Vebsite.
[0045]
"Percentage sequence klentity" het\veen two deoxyribonucleotide sequences
may also be detem1ined using the Kimura
2-parm1K~ter
distance model wfoch corrects fr)r
multiple hits, taking into account transitional and transversional substitufom rates, \vhile
assuming that the four nucleotide frequencies are the same and that rates of substitution do not
vaxy an10ng sites (Nei and Kmnar, 2000) as implemented in the lvlEGA 4 (Tmnura K, Dudley
13
WO 2016/054222
PCT/US2015/053239
J, Nd l'vl & Kumar S (2007) Jv1EGA4: Molecular Evolutionary
G1.~n1.~t1cs
Analysis (JVIEGA)
sofrware version 4.0. Molecular Biolo,·sY and Evolution 24:1596-1599), preferably vers1on
4.0.2 or later_
Tht~
gap opening and extension penalties arc set to 15 and 6.66 respectively.
Terminal gaps are not penalized. The delay divergent sequences s\vitch is set to 30. The
transition \Veight score is 35 set to 0.5, as a balance benveen a complete mismatch and a
matched pair score. The DNA \Ve.ight matrix used is the IUB scoring matrix where x's and n's
are matches to any lUB ambiguity symbol, and all matches score L9, and all mismatched
score 0,
[0046]
Pectin and
[0047]
The disclosed con1positions and methods include or utilize pectin or pectin-
Pectin~Related
Saccharides
derived sugars in order to sugars to enhance the efficacy of PGPR in regard to promoting
plant grmvth and plant health.
"Pectin" is a heteropolysaccharide found natively in the
primary cell \·Valls of terrestrial plants having a typical molecular \'i·'eight of 60··· 130,000 g/rnol,
wfoch varies based on the origin of the pectin and the extraction conditions. As used herein,
''pectin" is meant to include extracted pectin that has been extracted trorn its native condition
(e.g., extracted pectin from primary cell \'Valls of terrestrial plants).
[0048]
The pectin or pectin-related saccharides utilized in the disclosed composition
and methods may be isolated or substantially purified. The terms "isolated" or "substantially
purified" refers to pectin or pectin-related saccharides that have been removed from a natural
environment a11d have been isolated or separated, and are at least
6m"'o free,
preferably at least
75°/!1 free, and more preferably at least 90% free, even more preferably at least 950..{, free, and
most preferably at least 100% free from other components •vith which they \\'ere naturaHy
associated, \.Vhich other co1nponenis may include but are not limited to cellulose.
[0049]
Although the composition of pectin may vary among plants, pectin typically
has a composition in whkh D-ga!acturonic acid is the main monomeric constituent (i.e.,
typically D-galacturonic acid represents
>50~{,
of the monomeric constituents of pectin), The
D-galact.uronic residues of pectin optjonally inay he substituted \vi th D-xylose or D¥apiose to
14
WO 2016/054222
PCT/US2015/053239
fom1 xylogalacturomm and apiogalacturonan, respectively, branching from a D-ga!actunmic
acid residue.
So-called "rhamnogalcturonan pectins" contain a backbone of repeating
disacchmides of D-galacturonic acid and L-rhamuose.
[0050]
In nature, the majority of carboxyl groups of ga!acturonic acid in pectin are
esterified with methanol (i.e., >50Ch1 and as much as 800.1(i of the carboxy! groups of
galacruronic acid in pectin are esterified vvith methanol). During extraction, this percentage
may decrease \vhere ex.irncti<Jn inay result in hydrolysis of the ester bond, and extracted
pectins may be categorized as high-ester versus lmv-ester pectins having <50?0 of
galacturonic acid residues being esterified.
Non-esterified galacturonic acid units can be
either free acids (i.e., carboxyl groups) or salts with sodium, potassium, or calcium (i.e.,
galacturonaw salts).
[0051]
In nature, D-ga:lacturonic acid may be sy11thesized from D-gluconoric acid
derivafrves (e.g., from UDP-D-glucuronate via 4-epimerization) and conversely, Dgalacturonic acid in pectin may be metabolized to form D-gluconoric acid derivatives (e,g., 5dehydro-4-deoxy'-D-glucuronate v.ia ohgogalacturonate lysis). As used herein, pectin-related
saccharides include pectin-derived saccharides such as hydrolyzed pectin, D-galacturonic acid
(or D-galacturonate sails), and D-gluconoric add (or D-gluconorate salts), or combinations
thereof
[0052]
The compositions and methods disclosed herein may include or utilize a
saccharide that is a substrate for an enzyme or transporter encoded by a gene selected from
the group consisting of uxaA (a!trnnate dehydratase), uxaB (altmnate oxidoreductase}, uxaC
(uronate isomerase), uxuA (mannonate dehydratase), zcad? (D-rnannonate oxidoreductase).,
kc~gA
(4-hydroxy-2-oxoglutarate aldolase), kdgK (2-dehydro-3-deoxyg!uconokinase), exuR
(hexuronate utilizat1on operon transcdptional repressor), exuT (hexuronate transporter}, and
combinations thereof The compositions and methods disclosed herein rnay include or utilize
a saccharide that is a substrate for a pectinase enzyme (e.g,, an pectinase enzyme selected
from a gronp consisting of pectin lyase, p('Ctate !yase, polyga!acturonase, and pectin esterase).
15
WO 2016/054222
[0053]
PCT/US2015/053239
Substrates as such may include but are not limited to sat:charides derived from
pectin such as D-ga!acturonate and 0-g!ucuronate. The saccharide may comprise a mixture
of sugars or the saccharide may comprise a heteropolysaccharide. In embodiments in whkh
the
saccharide
lS
heteropolysaccharide,
a
heterogeneous
preferably
mixture
of sugars
D-galacturonate
or
monomenc
the
saccharide
units,
is
a
D-glucuronate
mcmorneric units, or the smn of D-ga!actun.mate monomeric units and D-g!ucuronate
monomeric units represent
>50~{i,
>60%, >7M·&,
>80~-'o, >90~··o,
or
>-95~··o
of total monomeric
units in the heterogeneous mixture of sugars or the heteropolysaccharide.
[0054]
The disclosed pectin and pectin-related substances may include synthetic
pectin. Synthetic pectin may include pectin synthesized by pol;'merizing pectin monomers
(e.g., uronic acid) in vitro to form pectin-like substance referred to as synthetk pectin. (See.
e.,g., U.S. Patent No. 2,156,223.
Fmtherrnore, the disclosed pectin and pecti1H·elated
substances may include naturally and non-natural1y occurring polyurnnic acids.
[0055]
Inoculants
[0056]
The presently disclosed PGPR may he formulated as an inoculant for a plant
The term "inoculant" means a preparntion that includes an isolated culture of a PGPR and
optionally a carrieL lnoculants comprising PGPR and carriers are known in the art. (See,
e.g., Bashan, "lnocnlants of Plant Gro\.vth-Promoting Bacteria for use in Agriculture,"
Biotechnology Advances, Vol. 16, No_ 4, pp. 729- 770, 1998)_
PGPR inoculants may be
administered to plants {e.g. to the roots of plants), to seeds (e.g., as a coating for the seed or at
the time that the seed is planted), or to soi I (e.g, to soil surrounding plants to be treated}
[0057]
A PGPR inoculant may be described as a fi_mnulation containing one or more
PRPR spedes il1 a carrier material, which may he an organjc carrier, an inorganic carrier, or a
cmrier synthesized from defined molecules. Optionally, the carrier may be sterile or sterilized
prior to be fonnulated \vith the PG-PR to form the PGPR inoculant Preferably, the carrier is
nontoxic, biodegradable and nonpolluting. In the disclosed inoculants comprising a pectin
16
WO 2016/054222
PCT/US2015/053239
saccharide, the pectin saccharide optionally may function as a carrwr or optionally the
inocu!ants may comprise a carrier other than the pectin saccharide.
[0058]
The carrier of the PCi:PR inoculant is the delivery· vehicle for the live PCiPR to
the plant, seeds, or soil. The carrier represent is the major portion by volume or weight of the
inocnlant. Suitable carriers may include liquids, po"\.vders
(e.g~,
having an average effoctive
particle diameter of 0J)75 to 0.25 nun), granulars (e.g., having an average effective particle
diameter of 0.35 to 1.18 nun), and slurries "\Vhich have the capacity to deliver a
sufficfr:~nt
number of viable PGPR. cells to tlle plant, seeds, or soil. Preferably, the carrier extends the
shelf-lite of the PGPR (e.g., such that the PGPR has a shelf-lite of at least I or 2 )"ears at room
temperature). Examples of carriers include but are not limited to peat, coal, days, inorganic
soil material, plant \Vaste materials, composts, fam1yard manure, soybean meal, soybean oil,
peanut oil, vd1eat bran, inert rnaterials such as vermiculite, perlite, phosphate, polyacrylarnide,
alginate beads, oil-dried bacteria. In some embodin1ents, the PGPR may be encapsulated by a
carrier, for example, \vhere the caJTjer is a carbohydrate that forms a matrix around the PGPR.
[0059]
The inoculant includes a suitable an1ot1nt of PGPR relative to carrier. In some
embodiments, the inoculant includes 102-lOn cfo PGPR per ml carrier (or per gram. can"ier),
or 104-10 10 cfu PGPR per ml carrier (or per gram carrier}, or 10 6 -10~ cfo PGPR per ml carrier
(or per gram carrier). The composition may include additional additives including buffering
agents, surfactants, adjuvants, or coating agents.
[0060]
The PGPR utilized in the disclosed composition and methods may be isolated
or substantially purified. The tenns '';jsolated" or "substantially purified" refers to PGPR that
have been removed from a natural environment and have been isolated or separated, and are
at least 6(YVii free, preferably at least 75'Vii free, and 1nore preferably at least 900.,.o free, even
more preferably at least 950.-'o free, and most preferably at least l 000...-;) free from other
components with \\'hich they were naturally associated.
An "isolated culture" refors to a
culture of the PGPR that does not include significant amounts of other materials such as other
materials "\vhich normally are found in soil in \Vhich the PGPR grov•ls and/or from ..-vhich the
PGPR nnrmaHy may be obta.ined.
An ''"isolated culture" rna,y be a culture that does not
17
WO 2016/054222
PCT/US2015/053239
include any other biological, microorganism, anzi.ior bacterial
spt~cies
in quantities sufficient
to intertere \.Vith the replication of the "'isolated culture." Isolated cultures of PGPR may be
combined to
prt~pare
a mixed culture of PGPR.
[0061]
J\,fothods of Treating Plants. Seeds, or Soil
[0062]
Also disclosed are methods of using pectin or pectin-related saccharides to
improve the efficacy of PGPR in regard to promoting growth or health in plants.
The
disclosed methods may include administering the above-described inocu1ants comprising a
PGPR and a pectin saccharide to plants, seeds, or sol!. In sorne embodiments, the disclosed
methods for improving plant gro\vth or plant health may include: fa) treating plants, seeds, or
soil \vith a plant grcnvth promoting rhizobacteria (PGPR) that expresses a protein associated
\'-'ith pectin metabolism and (b) treating the plants, seeds, or soil \.Vith a saccharide comprising
pectin or a pectin-related saccharide (e.g., hydrolyzed pectin, D-galacturnnate, D-glucuronate,
or mixtmes thereof), where the plants, seeds, or soil may he treated \Vith the PGPR and the
saccharide concurrently or in either order (i.e., the PGPR may be administered before,
concum.~ntly
•with, or after the saccharide is administered). The PGPR and pectin saccharide
may be formulated as an inoculant and administered concurrently to treat plants (e.g.,
administert~d
to the roots of plants), to seeds (e.g., as a coating for seeds), or to soil (e.g._, as a
soil amendment).
[0063]
The disclosed methods may be utilized to improve plant growth or plant health
by controlling soil-borne pests, Soil..:borne pests controlled by the d.isdosed methods may
include hut are not lilnited to nematodes and herbivorous insects. The disclosed 1nethods mav,·
be utilized to improve plant growth or plant health by controlling or treating a disease.
Disease controlled or treated by the disclosed n1ethods may include but are not limited to a
bacterial disease, a fungal disease, and a viral disease.
[0064]
The presently disclosed PGPR and pectin saccharide may he administered as
an inoculant for treating plants. The methods of t.reatmt~nt conternp!ated herein may include
treating a plant directly including ireating leaves, stems, or roots of the plant directly. The
18
WO 2016/054222
methods
(lf
treatment ccmteiriplated
PCT/US2015/053239
h(~rein
may include trnating seeds of tlw p!aut, e.g,
coating the seeds prior to the seeds being planted to produce a treated plant. The rnethods
contemplated herein also may include treating a p!aut indirectly, for exarnpk, by treating soil
or the environment surrounding the plant (e.g., in-furrmv granular or liquid applications),
Suitable methods of treatment may include applying an inoculant including the PGPR and the
saccharide via high or low pressure spraying, drenching, and/or injection. Plant seeds may be
treated by applying lo\v or high pressure spraying, coating, immersion, and/or injection. After
plant seeds have been thusly treated, the seeds may be planted and cultivated to produce
plants.
Plants propagated from such seeds may be further treated \vith one or more
applications. Suitable application concentrations may be determined empirically. In some
embodiments \'i,.here the PGPR and pectin saccharide are applied as a spray to plants, suitable
6
application concentrntions may include spraying 10 -10 18 colony tbrming units (cfu) per
hectare of plants, more com.monly Hf-10
15
cfu per hectare,
For coated seeds, in some
2
8
embodiments, suitable application concentrations may be bet\-veen l 0 - l0 cfo per seed,
4
7
preferably 10 -10 cfu per seed. In other embodiments, the PGPR ;md pectin saccharide may
2
be applied as a seedling root-dip or as a soil drench at a concentration of about 10 -10
' 1,
(>fu/m.
12
6
8
10'1- 1ow c f'.u/m.
' 1, or ab out 1c
, 1.
• > - 10 c t'wm
[0065]
l'vlethods of Treating Animals
[0066]
Also disclosed are methods of using pectin or pectin-related saccharides to
improve the efficacy of PGPR in rega.rd to prnrnoting grn\vth or health in animals.
The
disclosed methods may include administering the afore-described inoculants comprising a
PGPR and a pectin saccharide to anirnals (e.g., in the fcmn of an animal feed composition
such as a pelleted feed composition comprising the afore-described inoculants),
ln some
embodiments, the disclosed methods f()r improving animal e,-.rmvth or animal health rnay
include: (a) administering to an animal a plant grmvth promoting rhizobacteria (PGPR) that
expresses a protein associated with pectin metabolism and (b) administering to the anirna! a
pectin saccharide comprising pectin or a pectin-related saccharides (e.g,, hydrolyzed pectin,
D-galacturonate, D-glucuronate, or mixtures thereof), '.vhere the animals may he administered
19
WO 2016/054222
PCT/US2015/053239
the PGPR and the pectin saccharide concum.•ntly or in either order (i.e., the PGPR may be
administered before, concurrently with, or after the saccharide is administered).
[0067]
Feed compositions comprising the PGPR and pectin saccharide may be
administered to animals orally. Ora! administration includes, but is not limited to, delivery in
feed, 'Nater, by oral gavage or aerosol spray.
If supplied in an animal. feed, the feed may
9
comprise ben:veen l 0 4 and 10 cfu PG PR/gm of finished feed. Suitably the feed comprises
bef\veen 10 5 and 5 :< .10 cfu PGPR/gm feed. The PGPR and pectin saccharide ma~{ be added to
7
the feed during production, after production by the supplier, or hy the person feeding the
anirna1s, just prior to providing the food to the anin1a!s.
[0068]
The disclosed methods for promoting growth or health in animals may be
practiced in order to increase overall gastrointestinal health, improve production pedbnnance,
and reduce enteric bacterial pathogens of importance to both animal health and human food
safety. These PGPR and pectin saccharide may be added tn anin1a!. diets at the rate of about
4
10 to 1<P PGPR per gram of finished feed for optima! inclusion rate, if the bacteria or
probiotic compositions being administered continuously, and a higher inclusion rate may be
necessary if the PGPR or the compositions are provided intermittently. \Vhile administratlon
though the
ft~ed
is a preferred route of administration, the PGPR a11d pectin saccharide may
also he administered via drinking \vateL. through course spray, through aerosol spray, or
through a1Ty other means by which the agricultural animals may ingest these PGPR and pectin
sacdmride.
[0069]
JVlethods for Preparin_g the Disclosed Comr)Ositions and lnocu!ants
[0070]
Also disclosed are methods of using pectin or pectin-related saccharides tu
prepare compositions and inoculants as disclosed herein.
The methods may include
combining PGPR and plx:tin, \Vhich has b(•en extracted from pectin-containing plant material,
or pectin-related saccharides to prepare the disclosed compositions and inoculants.
Optionally, a caffier may be combined \Vith the PGPR and pectin or pecdn-re!ated
sacchaddes to prepare the disclosed co1npositions and inoculants.
20
WO 2016/054222
[0071]
PCT/US2015/053239
In some embodiments, th!;.~ methods may include combining l 02- ! 0 12 cfo PGPR
4
per ml carder (or per gram carrier), or I 0 -10
10
cfu PGPR per ml carrier (or per gram can'ier ),
or l 06-10 3 <.:ft1 PGPR per ml carrier (or per grain carrier). In some embodirnents, the methods
may include cornbining pectin, \.\/hi.ch has: been extracted frorn pectin-containing plant
material, or pectin-related saccharides may be combined \Vith PGPR and optionally a carrier
to prepare the disclosed compositions and inocu!.ants, \.vherein the pectin or pectin-related
saccharides are present in the prepared compositions and inoculants at a concentration of at
"'bc·,t1t
" 0'/ o,
te""t
'•" <»
' ' c·i· • 1~/
'0, 0 ·-·
0
1• . 0°/
/ 01· ~.
? 0°:/
'o, ·1....~ 'o,
. Q f,.,,fvt
\"
"
c1r \' 1/\t) to 'abot1t ()
'>.
.
~G/o,
, •. ,
l .()~/
5°/o,
'o, 1....
?
~. 0°/
'o,
or 5.0~/Q (v·i/\v or \v/v). ln some embodiments, the methods may include combining PGPR and
4
· at a concemrnt1on
· o f.· al)OUt
t.
t.
pectin
at 1east <wout
ll)~-, .to·~", l o ,
10
11
,
t 0 12, lOn, or 10
104, 105, 106,
HI
14
w-,
· -~
6
)9 H· ) 10,
l 0 , .to'·, i oj(·, ·It·,
3
cfo PGPR per gram pectin or pectin-related saccharides, to about 10
,
H/', 109 , 1010 , lOu, 10l2, 10u, 10 14, 10 15 cfu PGPR per gram pectin or
7
pectin-related saccharides (e.g., ranges such as 10 to l Ou cfu PGPR per grmn pectin or
pectin-related saccharides are contemplated herein).
In the rnethods,. additional additives
including buffering agents, surfactanis, adjuvants, and. coating agents 1nay be combined \Vlth
the PGPR, pectin or pecti1H·elated saccharides, and optional carrier in order to prepare the
disclosed compositions and inocnlants. Compositions and inocu!ants prepared hy the aforedisdosed methods also are contemplated herein.
EXAMPLES
[0071]
The folkJ\.ving Examples ;.u·e illustrative and are not intended to limit the scope
of the claimed subject matter.
Reference is made to Hossain
el
al., "Deciphering the
conserved genetic loci implicated in plant disease through cmnparative genomics of Bacillus
amyloliq11lfi:wiens subsp. plcmtarum strains,'' Frontiers in Plant Science,, 2015 Aug I 7;6:63 I
doi: 10.3389/tt)Js.2015JJ063L cCollection 2015, (hereinafter relerred to as "Hossain et al.,
Frontiers Plant Science 2015), the content of which is incorporated herein by reference in its
entirety,
[0072]
Example 1
~
Deciphering the conserved Qenetic loci implicated in plant
disease thrnu£h comparative genomics of Bacillus anv·to!iquefdciens subSJ3. plantarum strains
21
WO 2016/054222
PCT/US2015/053239
[0073]
Abstract
[0074]
Tu understand the grov·lth-promoting and disease-inhibit1ng activities of plant
growth-promoting rhizobacteria (PGPR) strains, the genomes of 12 Bacillus subtilis group
strains \Vith PGPR activity \.Vere sequenced and analyzed. These B. subtilis strains exhibited
high genomic diversity, \Vbereas the genomes of B. amylo!iq11E.fhciens st.rains {a member of
the B. subtilis group) are highly conserved. A pairwise BLASTp matrix revealed that gene
farnHy similarity among Bacillus genomes ranges from 32- 90%, \\ lt11 2,839 genes within the
1
core genome of /3. amyloliquef'aciens subsp. plantan1n1, Comparative genomic analyses of B.
arnyloliq1R:fc1ciens strains identified genes that are linked \Vith biological control and
colonization of roots and/or leaves, including 73 genes uniquely associated with subsp.
p!amarum strnins that have predicted functions related to signahng, transportation, secondary
metabolite
production~
and carbon source utihzMion. Although B. amyloliquej(:1ciens subsp.
plan/arum strains contain gene clusters that encode many different secondary metabolites,
only polyketide biosynthetic clusters that encode diffiddin and 1nacrolactin are conserved
within this subspecies. To evaluate their role in plant pathogen biocontrol, genes involved in
st.~condary
metabolite biosynthesis \Vere deleted in B. amyloliq1.1t:'.fdciens subsp. p!antarum
strain, revealing that difficidin expression is critical in reducing the severity of disease,. caused
by )tanlhomonas axonopodis pv. vesicatoria in tomato plants. This Example defines genomic
features of PGPR strains and links them with hiocontrol activity and \Vith host colonization.
[0075]
Introduction
[0076]
Bacteria associated with plant roots that exert beneficial effects on plant
gro,vth and develop1nent are refe1wd to as plant growth---promoting rhizobacteria (PGPR)
(Kloepper and Schroth, 1978;K1ocppcr et al., 2004), Bacillus and Pseudomonas spp. are
predominant among the diverse bacteria! genera that have been !inked -..vith PGPR activity
(Podile and Kishore, 2006), tvkmhcrs of the B. suhtilis group, including B. suhtilis, B.
lichentjbrmis, B. pumilus. B. amyloliquefaciens, 13. atrophaeus, B. mqjavensis, B. va!hrnwnis,
B. sonorensis, and B. tequilcnsis have been identified as PGPR strains for their capacity to
stimulate plant growth and suppress pathogens within rhizosphere and phytlosphere
22
WO 2016/054222
PCT/US2015/053239
(Kloepper et al., 2004;Hao et aL, 2012;Kim et al., 2012). Strains of B. arny!o!iqutfhciens
art~
\.Videly used for their positive effects on plant gnnvth (Idriss et aL, 2002). Reva et at (Reva et
al., 2004)
rt~port.ed
that seven Bacillus isolates from plants or soil are closely related yet
distinct from B. amyloliquef'ac:iens type strain DSJVlfr. In addition, these strains are more
proficient for rhizosphere colonization than other members of the B. subtilis group. GB03
(Nakkeeran et al., 2005} INR7
(Kokalis~Burd!e
et aL, 2002) and FZB42 (Chen el aL, 2007a}
are PGPR strains \Vithin the Bacillus subtilis group that have been \videly used in different
commercial formulations to promote plant gro'wth.
[0077]
In addition to promoting plant growth, PGPR strains tnay exhibit biological
contrnl of plant diseases. Antibiosis, through the production of inhibitory bioactive
compounds, and indtKed systemic resistance are widely reponed biological control
inechanisms of Bacillus spp. PGPR strains (Ryu et al., 2004). PGPR Bacillus spp. strains
produce diverse antimicrobial compounds including antibiotics (Emmert et at, 2004), volatile
organic con1pounds (VOCs) ('{uan et al., 2012), and lipopeptides (Ongena et aL, 2007) that
are associated with the observed biocontrol activity against plant pathogens. For example, B.
1.unyloliq1.u:fc1ciens NJN-6 produces 11 VOCs that provide antifungal
activity against
Fusarium O,\ysporum f sp, cubense (Yuan et aL, 2012). Similarly, B. sub!ils strains produce
tipopeptkks (e.g. surfactin and fongycin), that induce systemic resistance in bean plants
(Ongena et al., 2007). PGPR strains usually need to colonize plant roots extensively to exert
plant gro\vth promodng effocts using both direct and indirect mechanisrns (Lugtenberg and
Ka1nilova, 2009), extensive root colonization is not required for induced S)lSte1nic resistance
(JSR) (Kamilova et a!., 2005). In some PGPR strains, root colonization is a prerequisite for
biocontrol activit.Y through antibiosis (Chin et al, 2000). For example, H amyloliqmfaciens
subsp. plantmwn FZB42 exerts gr0\vtl1 promoting activities through efficient colonization of
plant roots (Fan et al., 20 t 1). Previously, it has been demonstrated that
over~expression
of
genes involved in phosphorylation of DegU, a hvo-cornponent response regulator of B.
an1yloliqw.:f'acie11s strain SQR9, positively influences root colonization as well as other
gmwth-promoting activities by PGPR strains for controlling cucurnber \'<lilt disease (Xu et al.,
20 ! 4 ). I\foreover, the root colonization capacity of a poor root colonizer can be improved by
23
WO 2016/054222
PCT/US2015/053239
cloning genes that are required for efficient mot colonization (Dekkers et aL, 2000).
Competitive root colonization by PGPR are controlled b:v many genes andior genetic
clustt~r(s)
(Dietel et al., 2013), so identification of these genetic loci involved in competitive
root colonization are challenging if genome sequences are Jacking for those PGPR strains
(Lugtenberg and Karnilova, 2009). Analysis of additional PGPR strains will help elucidate the
med11u11sms of competitive root colonization, antibiosis and JSR of PGPR strains and form a
foundation for genetic engineering and other strategies to increase the plant-growth promoting
capacity of these bacteria.
[0078]
In this Example, w·e sequenced the genomes of 12 Bacillus suhlilis group
isolates from diverse locales. Comparative genomic analyses of PGPR strains and control
strains of
tht~
B. subtilis group withuut m1y report<.":li biocontro! activity against plant
pathogens provides insight into genomic features involved in PGPR activity. PGPR strain
AP193, which inhibits grm.vth of plant and animal bacterial pathogens (Ran et al., 2012), is an
ideal candidate to evaluate the relative contribution of genes that me predicted to be involved
in the biosynthesis of bioactive secondary metabolites that could contribute to biocontrol
activity, specifically difficidin (lffnD mutant), surfoctin (sr/AA mutant), as \vell as all
polyketides and !ipopeptides produced by non-ribosomal peptide synthesis, including
difficidin (4jJ mutant), \fotants \vere then tested for their ability to inhibit plant pathogens in
vitro and control bacterial spot disease in tomato,
[0079]
~4aterials
[0080]
Bacterial strains, plasnrids and gro\\,.tb conditions.
and Methods
Bacterial strains and
plasmids used in this Example are listed in Table l. E. coli and Bacillus strains \·Vere grmm in
Luria-Bertani (LB) medium: hmvever, for electrocornpetent cell preparation, Bacillus
amyloliqu~1(lciens
suhsp. plantarum /\P193 was gro\vn in NCivl medium (17A g K::;HPO.h
11.6 g NaCl, 5 g glucose, .5 g tryptone, l g yeast extract, 0.3 g trisodium citrate, 0.05 g
!\..fgS(}r 7H20 and 9 L l g sorbito! in 1 L deionized \vater, pH 7. 2). For production of
secondary metabolites, Bacillus cultures \Vere gro\vn for 48 h at 3(Y"::C in Tryptic Soy broth
(TSB). In addition, ampkiUin (100 pgiml), chlormnphenicol ( 12.5 pgirnl) or erylhrornycin
24
WO 2016/054222
(200
~tg/ml
PCT/US2015/053239
frir L coli or
5~1g/ml
for Bacillus) were used as selective
ag,~nts
in grov·.rth media
as required.
[0081]
Sequencing, assemblv and annotation. Next-generation sequencing of Bacillus
spp. genomes v.:>as performed using Illumina and Roche 454 sequencing platfom1S. Indexed
l!lmnina libraries were prepared for sirains AP71, .AP79, and A.BO l using Nextern DNA
Sample Prep Kit (Epicentre, l\.1adison, \V!) and sequences were generated using an lllumina
l\!'liSeq with a 2 x 250 paired end sequencing kit Ba.rcoded lHumina libraries for strains
AP143, APl 93, and AP254 were constmcted using a NxSeq@ DNA Sarnple Prep Kit
(Lncigen, r....tiddleton, \iVl) and sequenced at EnGenCore (Univ. of South Carolina) using the
454-pyrosequencing platform. Genomic DNA library construction and sequencing for
Bacillus subtilis GB03, Bacillus pumilus INR7, H. nwiavensis KCTC 3706T, H. tequi!ensis
KCTC l3622T, Bacillus siamensis KCTC l3613T, and. B, .smwrensis KCTC l3918T were
conducted at the National lnstrnment Center for Environmental l'vlanagernent (Seoul,
Republic of Korea), using the Hlumina HiSeq 2000 sequencing platfonn. Sequence reads
'vere trimmed for quality then assembled de novo nsing the CLC Genomics Workbench
(CLCBio, Cambridge,
.~vlA )_
Gene prediction and annotation \Vere perfbnned using Genel\.fa.rk
(Lukashin and Borodovsky, 1998) and the RAST annotation server (Aziz et aL. 2008),
respt~ctively.
The identity of ind.ividual open reading frames (ORFs) frorn secondary
metabobte biosynthesis gene clusters \vas confirmed hy BLASTx against the GenBank
database.
( ··o
_1u{)"'
.>
Genomt~
sequence reads for strains ABOl, AP71, AP79, AP143, AP193, AP254,
1
• et a 1· ., ·-io14·
(·c·'_.1.101
,_ ' ),
IN·-1>
·"' -· 13·11 ·. '· ~/ {··1 eong et a1., "'0·14·
.:. · ), E:-,.7 C"-f'('
__ __ ,i7{)"··1··
..J _, KC"I-'C'
.. ___t .>6
_ (-. 1·eong et
al., 2012), KCTC 13918T, and KCTC !3622T \Vere deposited into the Short Read Archive
(SRA) at NCBJ under the accession numbers SRRl 176001, SRR1 l 76002, SRR l 176003,
SRRI 176004, SRRl 176085, and SRR l 176086, SRRl 034787, SRR 1141652, SRR 1 l 41654,.
SRR 1144835, SRRl l 44836, and SRR 'I 144837, respectively.
[0082]
Determination of average nucleotide identity. Average nucleotide identities
-
{AN[)- bet\veen genomes \\'ere calculated using an A.NI calculator that estirnates AN!
~
according to the methods described pre·viously (Goris et aL, 2007),
25
WO 2016/054222
[0083]
PCT/US2015/053239
Phvlogenetic anall'sis of Bacillus
SQ~ck~s.
For phylogenetic analysis, the gyrB
gene sequence for each strain (a list of the 25 strains is presented in Figure 1) \Vas retrieved
from sequence data. Strains AS43,3, FZB42, YAU B960l-Y2, CAll B946, and 5B6 ..-vere
used as representatlve strains of B. amyio/iqzu.fi.u::iens subsp.
and
TA208
plantarum~
\Vere used as representative strains of R
strains DSM?, LL3
amyloliqutfi.~cicns
subsp.
amyloliqut:J<.:1ciens. The gyrB phylogenetic tree \Vas inferTed with l'Vl EGA5 .05 (Tamura et al.,
2011) using Neighbor-Joining (Saitou and Nei, 1987) and Ivfaximmn Likelihood {fvIL)
methods (Felsenstein, 1981 ). All positions that co11tained gaps or missing data ..-vere
eliminated from the final dataset. resulting in 1911 bp positions of ,gyrB sequence. \Ve used
729,383 bp of DNA to represent the conserved core genome kmnd across 25 strains oft.he B.
sublilis group, to generate a phylogenornic tree using RAxrvlL ( v 7 2. 7) (Pfeiffer and
Stamatakis, 20 l 0). The phylogenomic tree \Vas then visualized \Vith iTOL (ltttu;L~i1£olLQIUQLgg)
(Letunic and Bork, 20.11).
[0084]
BLAST matrix.
The BLAST matrix algoriUun was used for pmrw1se
comparison of Bacillus PGPR strain proteomes, using r:nethods described previously (Friis et
al., 2010). The BLAST matrix detem1ines the average pen.:ent s1n1ilar1ty between proteomes
hy measuring the ratio of conserved gene families shared between strains to the total number
of gene familit$ within each strain. The absolute number of shared and combined gene
families for each strain \vas displayed in matrix output This matrix shmvs the number of
proteins shared benveen each proteome.
[0085]
Corc-g:cnome analvsis.
The core-genome of 13 Bacillus spp. strains \Vas
generated using coding and non-cod.ing sequences. \Vhole genmne sequences fron1 these
strains were aligned using progressive JVlauve (Darling et al., 2004), \\ hich identifies and
1
aligns locally collinear blocks (LCBs) in the XlViFA fi)nnaL LCBs from alignments \Vere
collected
us mg
strip Subset LCBs
{J1!J;ii;L'.g~1Jt!m.h;L~~:i~iLJ;_~it1Lm_mnJ,~l~m112B.hf21fu".),
us mg
minimum kne.rt.hs of 500bp. AH LCBs were concatenated and converted to nrultifasta format
using a per! script. The same protocol \Vas used to obtain all core sequences, with the
exception that the minimum lengths of LCBs \>i-'ere 50 bp, instead of 500 bp. The Bacillus spp.
26
WO 2016/054222
PCT/US2015/053239
core genome was obtained from the comparative allgnment of a!l compkte Bacillus spp.
genomes available in the OenBank as of August 2014 (n=81 genomes). The core genome of
tht~
B. sublilis group \Vas obtained from comparative analysis of 53 "\Vho!e genomes of B.
subtilis strains that included 41 genomes obtained from GenBank and 12 PGPR genomes
sequenced in this Example. B. amyloliquefaciens species-level and B. amyloliq1.u;laciens
subsp. plan/arum-level core genomes \Vere generaied from 32 B. amyloliquefctciens and 28
subsp. pion/arum genomes. Core genomes \Vere exported to the CLC Genomics Workbench
(v 4.9} for evaluation of alignments and annotation using the RAST server (Aziz el al., 2008)_
The hst of Bacillus spp. strains used for core genome determination is provided in Table ? .
Additionally, to identity GPR-spedfic core genes, ra\\' sequence reads of PGPR strains
sequenced in this Example vvere sequentially reference rnapped against the genome sequence
of non-PGPR strain B. subtilis subsp. sufuilis str. 168
a(~cording
Lo methods described
previously (Hossain et al., 2013}.
[0086]
Identlfication of core genes uniguelv present in B. <.mu!foliqut;{ju:iens subsp.
plantarum strains. The aligned genome sequences of 32 11. amy!oliqw,faciens strains and 28
B. amyloliquefc1ciens subsp, plantarum strains (\·Vhich \i,.rere included within the B.
amylo!iquefaciens strains) \vere analyzed using CLC Genomics Workbench to obtain the
respt~ctive
species- and subsp, -level core genomes. Trimmed sequence reads of subsp.
plan/arum strain i\Pl 93 \Vere reference mapped against the subsp. plantarwn core genorne to
obtain
cort~
genome-specific sequence reads. The parameters of reference mapping were as
follows: xnisnrntch cost ===2,. insertion cost ===3,. deletion cost ===3. length fraction === 0.5,. and
~
similarity
= 0.8. Sequence reads mapped
~
to the subsp, plamarwn core genome were then
mapped against the species amyloliqut.:fi.1ciens core genome
w obtain unmapped sequence
reads. These unmapped sequence reads, represent the subsp, plantarum core genome that is
absent in the arnyloliq11(/i1ciens species-kvel
<.:on.~
genome, \Vere assembled de novo using
CLC Genomics Workbench then the resulting contigs were uploaded to RAST for gene
prediction and annotation. Each ORF, exclusively encoded by the plantarum core genome,
,.vas fortl1er confinned for uniqueness using BLASTn analysis against the genome sequences
27
WO 2016/054222
PCT/US2015/053239
of 28 B. amyluliqu,:.A:tciens subsp. plan/arum and four B, c.nny!o!iquej;:1ciens subsp.
amyloliquefi.Kiens strains listed in Supplementary Table L
[0087]
Prediction of secondarv metabolite biosvnthesis gene clusters in PGPR strain
AP 193.
Secondary metabolite biosynthesis gene clusters for strain AP I. 93 \.Vere predicted
using the secorn.iary metabolite identification tool anti SM.ASH (Bhn et al., 2013).
Pri.rner~
\Valking PCR \Vas used to fill gaps between contigs containing gene dusters encoding
secondary metabolite biosy11thesis. Gene prediction and annotation \.>./ere can"ied out hy
Gene!vtark (Luka.shin and Borodovsky, 1998) and BLASTx (NCB1), respectively.
[0088]
DNA manipulation and plasmid construction for PGPR strain /\PJ93
mutagenesis. Chromosomal DNA was isolated \Vith the E.L N.A. Bacterial DNA Isolation
Kit (Omega BioteL. Atlanta, GA) and plasmids \vere isolated •vith the E.Z.N.A. Plasmids
l'vfini Kit U (Omega Biotek). Primers used in this Example are listed in Table 2. Gene deletion
constmcts were assembled using splicing through overlap extension PCR (Horton et aL,
I 989). The assembled products •vere gel purified \Vith Gel/PCR DNA Fragments Extraction
Kit (JBl), digested \Vith appropriate restTiction enzymes, and doned into a pNZTl vector to
construct the delivery plasmids for gene replacement
[0089]
Jn
vi1ro
plasmid
methv!ation
usilrn
cell
free
extract
of
Baci!ius
arnvlolique/C1ciens subsp. p/antarum AP"! 93. To rnethyla.te plasmids prior to transformation
into 8- arnJ·!oliqu(faciens suhsp. plantarum AP19J, ihe method developed for Lactobacillus
plantarum \Vas used \Vith minor modifications (Alegre et at, 2004). Cells from a l 00 ml
overnight culture of strain AP193
(ODr;1~J
===
1.3-1.5) were pelleted hy centrifogation (8000
xg). washed \Vith 100 ml of chilled PENP but1er (10 n1ivI potassium phosphate, 10 111iv1
EDTA, 50 mM NaCl and 0.2 mM
P~ASF,
pH 7.0), and then re¥suspended to a final vo!mne of
4 ml. Cells \Vere disrupted by performing t\vo bursts (amplitude 50, pulse 3 and vrntts 25<30)
for 5 n1in each \vith a pause of 2 min, using a Vibra-Cell sonicator, and cooled \Vith ice to
prevent overheating. Cell debris was removed by centrifogation (8000 x g) at 4"'C and the
extract \Vas <.:ollected through decanting. Three ml aliquots of extract 1vere mixed \vith 3 ml of
glycerol ( 1OO~··o v/v) and 0.6 rnl of bovine serum albmnin ( l tng/ml), then stored at -20''C.
28
WO 2016/054222
[0090]
PCT/US2015/053239
The DNA modification assay \Vas perfom1ed in a final volume of 100 pl of the
following: 53 ~d TNE buffer [50 mi\.·1 Tris (pH 75), 50 mivf NaCL lO m~vl EDT/\], JO ~ti Sadenos·ylmethionine (0.8 ml'vi), 2
~ti
BSA (5 mg/ml), 25
~tl
cell free extract derived from strain
AP 193 and 10 p.I plas1nid DN/\ extracted from E. coli K 12 ER2925 (0. 5-l pg/pl}. The
mixture \:vas incubated at 3r··c for 16 h. :\kthylated DNA \Vas extracted ·with a DNA Clean &
Concentrator Kit (Zymo Research, CA}, then re-suspended in \Vater and stored at -20'"C.
[0091]
ElectrQtransfiJrmation of B. amvlolique{aciens suhsp. pianrarmn .AP 193. For
preparation of electrocompetent cells, strain AP193 \vas grown overnight in TSB, then diluted
100-fold in
NC~4
to inoculate a subculture. The culture was grmvn at 3T'C on a rotary shaker
until the OD600 reached 0.7. The cell culture \vas cooled on ice for l 5 n:1in and subjected to
centrifi.1gation at 8000 x g for 5 min at 4c·e, After \vashing four times \Vith ice cold ETM
buffer (0-5 l'vl sorbitol, 0.5 M ma1mitol, and JQO,.·~i glycerol}, electrncmnpetent cells \Vere resuspended in l.ilOO volume of the origina.J culture (Zhang et at, 2011). For electroporation,
J00 pJ of cells \Vere mixed
\'iii th
JOO ng of plasmid DNA in an ice-cold eledroporalion
cuvette (l mm electrode gap). Cells vvere exposed to a single 2 l kV/cm pulse generated by
Gene-Pulser (Bio-Rad Laboratories) with the resistance and (:apacitance set as 200 Q and 3
~tF,
respectively. The cells \.Vere immediately diluted into l ml of recovery medium (NCl1v1
plus 0..38l'vl mannitol) (Zhang et al., 201 l) and shaken gently at 30°C or
3r·c for 3 h to allow
expression of the antibiotk resistance genes. Aliquots of the recovery culture \vere then
spread onto LB agar supplemented \vith appropriate antibiotics.
[0092]
Tw·o-step replacement recombination procedure for the modification of the
strain AP193 genon1e. A two-step replacement recon1binationwas perfonned as previously
described, \vith minor modifications (Zakataeva et aL, 2010). To integrate the plasmid into
AP 193 's chromosome, a single crossover between the target gene and the homologous
sequence on the plasmid must occur. To do this, AP193 that contained a delivery plasmid
'\Vith theddetion constnKt 1.-vas first grmvn in LB broth for 24 h at 37'-'C (a non-permissive
temperature for plasmid replication}. Next,. the culture was serially diluted, plated onto LB
agar plates \Vitl1 erythromycin, and incubated at 3r''C. Clones \vere screened by colony PCR
29
WO 2016/054222
PCT/US2015/053239
using nvo sets of primers. Each set of primers aimeals st;quences specific to one of the
homologous fragments and to the chromosomal region just outside of the other homologous
fragmt~nt.
If PCR products had a reduced size, relative to the \.vi!d-type genotype for either
primer set. this indicated successful chromosomal integration of the plasmid. In the second
step, clones of the integrant \Vere cultured \Vlth aeration in LB at
3<J~c
for 24-48 h to initiate
the seccmd single-crossover event, resulting in excision of ihe plasmid, yielding erythrornycin
sensitive (ErnS) clones \Vith either a parental or a mutant allele on the chromosome. Colony
PCR was used to examine the presence of desired mutations by primer sets that flank the
deleted sequence.
[0093]
Constmction of strain AP 19 3 mutants defective in secondarv metabolite
biosynthesis.
AH mutant st.rains generated in this Exaniple are indicated in Table 1. The
disruption of the dfnD gene was achieved as follov,.'s: DNA f:ragrnents corresponding to
positions -867 to +247 and +643 to + 1570 \.Vith respect to the l{fhD translation initiation site
\·Vere PCR amplified using AP193 genornic DNA as a ternplate. The 1.\VO fragments \Vere then
assembled hy fusion PCR. A frameshift mutation \vas introduced during fusion to ensure
complete disruption of the gene. The deletion construct \Vas digested with XhoI and Spel,
then cloned into pNZTl ,. yielding pNZ-dif The plasmid was methylated in vitro as described
above and introdtKed into strain API 93 by electroporation. Once introduced into strain
AP I 93, plasmid pNZ-dif generated the 1sogenic mutant AP l 931\dfitD by tv,..-o-step
replacemt~nt
[0094]
m;.:ombination.
To generate the
,~/]1
deletion mutant, DNA fragments corresponding to
positions -781 to +29~ with respeci to ihe ,sjj) translation initiation site, and +95 to+ 935, \vltJ1
respect to the 4jJ translation tem1ination site, \Vere PCR ar:nplified using AP193 genon:ric
DNA as template, assembled by fusion PCR, digested 'vith Hindlll and Pstl, and cloned int.o
pNZT l to construct pNZ-sfp. The plasmid pNZ-sfp was used to generate rnutant AP l 93A4}J
using procedures described above.
[0095]
The A~1fAA mutant was obtained as folknvs: DNA fragments corresponding to
positions +5375 to +6091 and +6627 to +7366, ,.,.,.'iih respect to the s1:1AA translation initbtion
30
WO 2016/054222
PCT/US2015/053239
site, \Vere ?CR-amplified, fused by fusion PCR, digested whh HindUI and Pstl and cloned
into pNZTl as pNZ-srf Similarly, a frameshift mutation \vas introduced during the ftision of
tht~
upstream and do\vnstream fragments of the target deletion sequence to ensure complete
disruption of the gene. The plasmid pNZ-srf \\'as used to generate mutant AP l 93,\stfAA using
procedures described above.
[0096]
In vilro antimicrobial activities of PGPR strain AP 1.93 and its mutants .:urainst
plant pathou.ens. Plant pathogens Pseudomonas .\Jringe pv. tahaci, Rhizobium radiobacter,
)(anthomonas axonopodis pv. vesicatoria,. and ){anthomona8 axonopodis pv. campestris \vere
grov/n in TSB until the OD{;oo reached LO. The \vild tyJ)C strain AP193, as \vdl as the three
isogenic mutants A<{fhD, /1.~fj.J, and l\s1./AA developed in this Example, \Vere gro\v11 at 30''.·c in
TSB for 48 h at 220 rpm. Cultures \Vere then centrifuged at I 0,000
supernatant was passed through a 0.2
f1Hl
;v:
g for 2 min then
nylon filter (V\VR, PA). For antihiosis assays, 100
p.I of an overnight culture for each plant pathogen •vas spread onto TSA plates (Thenno
Scientific, N'{) separately then sted Ie cork borers ( l 0 mm diameter) \Vere used to bore \Ve lls
in agar plates. Filtered supernatant of AP 193 and its three nmtants \vere separately added to
fill \veils. Plates \),.'ere aHc.nved to dry and then incubated at 30°C overnight Zones of
inhibition \Vere measured and compared bet\veen mutants and wild-type strain /\Pl 93 to
determine tht~ir ant.in1icrobial activities against plant pathogens.
[0097]
LC-!v1S analvsis of bacterial supernatants, Bacteria! cultures were grmvr1 in 2
inl TSB for 72 hours and then cells were ren1oved by centrifugation at l 0, 000 x g for 10 min,
foHovled by 0,2
~ml
filtration of the culture supernatant. Samples were analyzed by direct
injection from m/z 50-1200 on a ultra-high pressure Liquid chromatography/QTof'.-mass
spectrometer (\Vaters Acquity UPLC and Q-Tof Premier, tviilford, 1'v1A) operated at a spray
voltage of 3. 03 kv and the source temperature of l OtY'C, The !'v1S analysis \Vas conducted in
negative io11 mode with a nwbile phase of 95(!/o acetonitrile, 50.··o \.vater and 0.1 (~lo formic ac.id.
[0098]
In vivo antibiosis of strain AP 193 and its rnntants a!.!a.inst a plant pathogen.
Rutgers tomato seeds (Park Seed, USA) \Vere
S0\\.'11
in Styrofoam trnys. Three weeks after
planting, seedlings were transplanted into a 4.5 inch square pot \Vith cornmercia! potting
31
WO 2016/054222
substnm.~
PCT/US2015/053239
(Sunshine mix, Sun Gro Horticulture, Agav>'am, 1'v1aine). Three days aft.er
transplanting, plants were sprayed \vi th sterile water or PGPR eel I suspensions ( J0 6 CFU/ml)
that had been \Vashed three times prior to being resuspended in stcrlle \Vater and nom1a1ized at
an ODMHJ = LO before being serially dilute& PGPR-inoculated plants were placed into a de\v
chamber at l 00%1 humidity in the dark for two days at 24'T then transferred to the
greenhouse. One day later, plants \Vere chaHenge-inoculated with
x:
axmwpodis pv.
7
vesicatoria by spraying approximately 10 ml of a 10 CFU/ml pathogen suspension over each
plant. Pathogen-inoculated plants were placed in the dew· chamber for t\vo days then placed in
the greenhouse. Plants \Vere watered once daily, Disease severity ratings and harvest \Vere
conducted after 14 days of diallenge-irmculation. For disease severity rating, four compound
leafo:; \Vere selected from the bottom of each plant The disease severity of each of the
cornpound leaves was dctennincd by rating the dis'-~asc severity of each leaflet and calculating
the average rating for the cmnpound leaf. Leaflets "vere rated using a 0-4 rating scale, \Vhere
O=healthy lea.flet, l= <20?.··(, necrotic area of the leaflet, 2= 20-500./Q necrotic area of the leaflet,
3==== 51-80 0,.(, necrotic area of the leatlei, 4=== 80-100 % necrotic area of the leaflet. ln addition,
dry shoot and root \Veights \Vere determined. The experirnenta1 design \vas a randomized
complete block \1>-'tth ten replications per treatment. The experiment \.vas conducted t>vice.
[0099]
Data analysis. All data wen.' a11alyzed by an analysis of variance (ANO VA),
and the treatment means \Vere separated by using fisher's protected least significant
diffort~nce
(LSD) test at P=0<05 using SAS 9.3 (SAS Institute, Gary, NC, USA).
[00100]
Results
[00101]
Genome Statistics and genetic relatedness of Bacillus spenes.
Genome
sequences of 12 different PGPR Bacillus spp_ strains were determined using next-generation
sequencing. The summary statistics for each Bacillus spp. genome sequences and their
assemblies are presented in Table 2. The approxi1nate sizes of Bacillus spp. genomes ranged
from 2.95-4.43 \.fop with an average genome size of 3.93 Mbp, \vhic.h is similar to the 4.09
l\ifbp average
gt~nome
size of complete B. subti!is genomes available in GenBank (April,
2015). The percent G+C content of the 12 PGPR Bacillus spp. strains ranged fron1 41.3-
32
WO 2016/054222
PCT/US2015/053239
46.6%, averaging 45.15°.{i , \Vhich is similar to the average percent G+C content of the B.
subtilis genome sequences ava1!able in GenBank (43. 72~·(,) (Ivtarch, 20 l 5 ). Painvise average
nucleotide klentltk$ (ANl), a newly proposed standard for species definition in prokaryotes
(Richter and RosseH6-JV16ra, 2009), were calculated for 13 Haci/lus PGPR strains to
determine their interspecies relatedness among Bacillus species. The ANI 'Values fiJr PGPR
Bacillus spp.
st.rains ABOI, AP71, AP79, AP!43, AP!93, and GB03 against B.
atnJ1oliq1w/aciens FZB42 (Chen et aL 2007a) \Vere greater than 98'}(i (data not shown),.
indicating that these PGPR strains are affiliated \Vith the B. arnyloliqu(:filciens species. The
98.88~/Q
ANI of PGPR strain AP254 to B. subtilis suhsp. suhtilis strain 168 suggests that
AP254 is affiliated \1>-'ith B. subti!is (data not shtw.m). The pain.vise ANI comparison of PGPR
.
r·N'R
strams
· . ...,i, K"('"'"'('('"'
· .. .
otlH.~r
"'7{
.,.I,,
J·
1 )6.
·K··c··1·c·
· . .
t'"l
..
1 _,()
l"'J'
,) .. , K···c'·l'('
·
.·.
.·
j""'(
.;) 18''.l'
.· . ,
1·">,C'J·!"I•
•
an. d.K.("["('
. . ....
)O..:..-.
aga.mst
eac11
produce ANI values kss than 95°10 (data not sho\vn) suggests that they are distantly
related to each other and represent diverse Bacillus species.
[00102]
Phylogenetic relationship of Bacillus st.rains. A phylogenetic mrn1ysis based
on gyrB gene sequences shmved sufficient resolution among Bacillus taxa and \vas consistent
\\iith ANI comparisons. Strains AP71, AP79, AP143, AP193., ABO!, and GB03 \Vere grouped
togetl1er \Vith reference strains of R amyloliquej(wiens suhsp. plantarum \.Vith h1gh bootstrap
support, indicating thm they are affiliated \Vith subsp. p!antamm. The three strains of B.
amyloliquefi1ciens subsp. amy!oliquefaciens DSM?, TA208, and LL3 clustered as a single
clade, separated from strains of subsp. plantarum, supporting the division of t\vo subspecies
in B. amy!o/iqw.j(.Kiens (Borr1ss et al., 201 l). The placement of strain AP254 with B. subtilis
subsp. subtilis strain 168 as a single clade \l.tith strong bootstrap support suggests its affiliation
ivvith members of the B. subtilis group (Fig. lA) ..A gyr!J gene based phyl.ogenetic tree
constructed using J\faximum Likelihood (rvlL} methods was also concordant with the
phylogeny constructed using Neighbor-Joining methods (data not sho•vn). ln addition to the
gyrB-based phylogeny, \.Ve constmcted a phylogenomic tree using 729,383 bp of core genorne
sequences present within the genome of 25 H. subtilis group isolates to provide a more refined
phylogenetic place1nent of PGPR strains. The topology· and allocation of strains to clades in
the gyrlJ phylogeny \Vas similar to the phylogenomic tree (Fig. 1B). One notable difference is
33
WO 2016/054222
PCT/US2015/053239
that the wpology of the tree regarding the position of strain B. siamensis KCTC 13(')'! 3 diffhs
significantly bet\veen the gyrB-based tree and the phylogenomic tree, \vith the gyrB based
phylogeny placing KCTC 13613 in a separate dade whereas the phylogenomic tree included it
\Vi thin
a monoph;,!etic group that includes strains of J{ amyloliquefi.:tciens subsp. p!antarum.
[00103]
BLAST matrix. Genom.e \Vic.le proteome comparisons of 13 PGPR Bacillus
strains using an all-against-all BLASTp approach demonstrated that PGPR Bacillus spp.
stTains are highly diverse, as indicated by gene family similarity betv./een PGPR Bacillus spp.
genomes ranging from 32-900,·(i (data not show·n). Consistent ,,.,..·ith the phylogenetic analysis,
high similarity was found among strains AP71, AP79, APl93, ABOl, GB03, and FZB42,
i.vith prnteomic similarity ranging from
[00104]
progressive
Core-a.enome analysis.
~fauve
70-90~··0.
Analysis of genome sequence alignment usmg
determined that the core genome of 13 PGPR Bacillus spp. strains
contains 1,407,980 bp of genomic DNA which encode 1,454 ORFs (data not shown).
Comparison of core genome sequences of the genus Bacillus, subgroup B. subtilis, species B.
amyloliqut.:f<.:1ciens, and subspecies plantarum demonstrated that as the number of genomes
increases,. the number of d1ft'erent subsystems \Vithin each respective core genome decreases
(Fig. 2A-C).. The highest mm1bers of subsystems in each of the core genome categories,
except for the genus Bacillus core genome, was devoted to carbohydrate metabolism. These
findings suggest that strains from the genus Bacillus nse diverse carbon smirct$. [n additiou,
the core genome for the genus Bacillus has rnore subsysiems devoted to RNA, DNA, and
protein metabolism compared to carbohydrate metabolism (Fig. 2A-C).
[00105]
The genome alignment from 28 different subsp. plantarum strains, including
six subsp. plantarum strains sequenced in this Exmnp!e, identified 2,550,854 bp of core
genome sequence that is predicted to encode 2,839 ORFs. The genome alignment of 32 B.
{.nnyloliqlH;'.fiJciens strains, including 28 subsp. plantarum strains, idendfied 2,418,042 bp of
core genome sequence predicted to encode 2,773 ORFs.
34
WO 2016/054222
[00106]
PCT/US2015/053239
The genome alignment of 53 strains of H. suhlilis group, including the I 2
strains sequenced in this Example, identified 578,872 bp of core genome sequence predicted
to
encodt~ 674 ORFs.
The number of protein coding genes present w1thin the genome of
Bacillus spp. (--4,000J and the lmv m1mber of ORFs (674) encoded by their core genon1es
suggests a large amount of genomic plasticity among Bacillus genomes that experience
frequent gene acquisitions and losses. It \·Vas observed ihat ihe 8. amyloliqut.;f(..tciens core
genome \Vas devoid of mobile genetic elements, such as prophages, transposable elements,
and plasmids (data not shc.rwn). Furthermore, the B. suhti/is core genome \Vas also devoid of
genes or genetic clusters !inked \Vith iron acquisition and metabolism, secondary metabolite
biosynthesis, si6">1lal transduction and phosphorus
[00107]
rm.~tabolism
(Fig. 2A-C).
In this Example, the genus Bacillus core genome v•las also determined by
analyzing all complete genorne sequences frmn the genns Bacillus cunently available in
GenBank. \Ve determined that the genus Bacillus contains 194,686 bp of core sequence
predicted to encode 201 different ORFs. The predicted functions present in all Bacillus strains
are limited to the following subsystem foatures: cofactor synthesis,, vitamin synthesis,
prosthetic groups and pigments biogenesis, cell \Vall and capsule biogent.$iS, membrane
transport, RNA. metabolism,. nucleoside metabolism, protein metabolism, regulation and cell
signaling, DNA metabolism, respiration, amino acids and derivatives, sulfor metabolism, and
carbohydrate utilization
[00108]
Comparative analysis of core genes unique I)'' present in B, mrwloliquqfctciens
subsp. p/antarum. Comparison of PG PR-specific genomes "vith that of non-PGPR B, subtilis
subsp. subtilis str. 168 did not identify any genes other than essential housekeeping genes that
were conserved \Vithin the genomes of PGPR strains (data not shnw11). Comparative analysis
of core genomes from 28 B. amy!oli.quef{:1ciens subsp. plamamm and 32 B, amyloliquf;/f:.1ciens
species identified 193,952 bp of sequences that are present vvithin the suhsp. p!anlarum core
genome but absent in the B. amyloliqmf1.1ciens core genorne. Among these genetic loci there
\vere 73 genes shared by all 28 plantarum strains but \vere not present in any strains of subsp.
amy!o!iqmfaciens, The putative functions of these genes includes transportation (7 genes),
35
WO 2016/054222
PCT/US2015/053239
regulation (7 gent$), signaling (l
gt~ne),
carbon degradation (I 0 genes), synthesis of
secondary metabolites (19 genes), and hypothetical proteins (12 genes) (Fig . 2C). Sorne of
tht$e
gt~ne
products may be involved in inwract1ons \Vith plants and rhizosphere competence
of subsp. plantarum strains (e.g., pectin utilization). For instance, genes required for uptake
and use of D-galacturonate and D-glucuronate are shared among genomes of B.
amyloliqu(~fiiciens
subsp. plantarum strains.
These include uxuA (mannonate dehydratase
(EC 42.1.8)), kdgA (4-hydroxy-2-oxoglutarate aldolase (EC 4. L3J 6)), k{}gK {2-dehydro-3deoxygluconate kinase (EC 2.7.1.45)), exuT (hexuronate transporter), exuR (hexurnnate
utilization operon transcriptional repressor), and uxuB (D-mannonate oxidoreductase (EC
1. l .1.57)).
In addition, genes required for biosy11thesis of the polyketides difficidin and
macrolactin v•iere consistently found in PGPR suhsp. plamarum strains, suggesting their
relevance in the biocontrol activities of these strains.
[00109]
Gene clusters encodinu secondarv metabolite biosynthesis and natural
competencv in strain AP 193.
Due to our observations of beneficial interactions betvveen
PGPR strain AP 193 and both plant and animal hosts (Ran et al., 2012), we selected this strain
fbr more intensive genome analysis. Assembly of strain AP193 genome sequences de nm'o
resulted in l 52 contigs larger than 1 kb, with a combined length of 4, 121,826 bp. Analysis of
AP 193 contig sequenct$, using tl1e antiSlV1ASH secondary metabolite prediction program,
suggests that gene clusters were present that are responsible for synthesis of three different
polyketides: bacitlaene, macrolactin and dif1icid.i11. In order to provide cornplete sequences for
these biosynthesis path\vays, the gaps hei\veen contigs 5 and 6, contigs 33 and 38, as v..·el! as
contigs 27 and 28 \Vere filled using PCR, foUo,ved by DNA sequencing. Each of the gene
clusters in A Pl 93 are collinear to their counterparts in B. cany!oliquej(1ciens
FZB42~
a
naturally competent plant root-colonizing B. c.uny!oliqz((Efi.Jciens isolate \vith the ability to
promote plant growth and suppress plant pathogens (Chen et aL, 2007a), The percent amino
acid identities of the proteins encoded by those clusters \Vere . . . . . ithin the range of 98-1 OO(~·o
when compared with those of FZB4L Secondary metabolite biosynthesis gene dusters
involved in non-ribosomal synthesis of cyclic lipopeptides surfactins, fengydn and
bacillomycin D and of the antimicrobial dipeptide bacilysin present in FZB42 were also
36
WO 2016/054222
PCT/US2015/053239
detected in the AP193 genome. The
perct~nt
amino acid id.entities of the AP193 proteins
encoded on those clusters to the FZB42 homologs ranged from 980.l!i to l 000.·o. The lack of
natural
compt~tency
of the PGPR strain AP 193 prompted us to detem1ine the presence of
competence-related genes \.Vithin this strain. We searched the AP 193 genome sequences for
the presence of competence related genes fbnnd \Vithin the genome of FZB42, and observed
that all of the genes required for encoding the structural. cornponents of the competence
system found in strain FZB42 are present \vi thin the genome of AP 193 \.vi th 98 to 1000...-o
identity (data not shmvn); however, genes comQ, comX, and comP are involved in regulating
quorum-sensing in H mnyloliquejiu:iens FZB42 (Chen et aL 2007a) \Vere absent \Vithin the
genome of strain APl93 (data not shO\-vn). The absence of comQ, com){, and comP may be
responsible for the lack of natural competency for strain AP l 93,
[00110]
API 93 secondarv metabolites inhibit the grmvth of multiple bacterial plant
pathogens in vitro. Antimicrobial activities of strain .AP 193 and its nmtants AP 193!\if,fhD
(deficient in the production of difficidin), APl 93i\srfAA (deficient in surfactin production),
and AP l 93/\.~_fj) (unable to produce polyketide or lipopepetide due to a deletion of sfj> gene
encoding
4'-phosphopantetheinyl
transferas!;.~}
\.Vere
tested
against
Pseudornonas ,):yringe pv. tabac.i, Rhizobiwn radiobacter, Xanthmnonas
vesic.atoria, and Xmtlwmonas axonopodis pv,
campt~stris.
demonstrated strong antimicrobial activity, \vhereas the
inhibitory
effi.~ct
plant
pathogens
axonopodl~
pv,
The AP 193 \vi!d type strai11
AP193A~'.fj>
mutant \vas devoid of an
against those plant pathogens (Fig. 3), underlining the contribution of
lipopeptides and/or polyket.ides in the hioactivity of AP193. This also indicates that the
d.ipeptide bacilysin, \.Vhose synthesis is independent of Sfp, was not involved in antagonistic
activity expressed in ·vitro. The AP193..'.\!uf4A mutant conferred antimicrobial activity similar
to \vild-type to P ..\)Tinge pv, tabaci, R. radiobacter, X axonopodis pv. vesicatoria, and J:.~
axonopodis pv. campestris (Fig. 3), suggesting that surfa.ctin has no putative role in the
antibacterial activity of AP193 against ti1ose plant pathogens under the conditions tested in
this Example. These findings also demonstrated that surfactin neither influences the
antirnicrnhial cornpound biosynthesis in AP 193 nor does it inhibit antibacterial activities of
the antibacterial compounds produced by APl 93. Difficidin acts as the im\jor antibiotic in
37
WO 2016/054222
PCT/US2015/053239
antagonism of A.Pl 93 against plant pathogens P.
,~yringe
pv. tabaci, R. radiobacter, )(
fft.:onopodis pv. vesicatoda, and .X ax:onopodis pv. campestris as indicated by the lack of the
inhibitory effb;.t of tl1e AP193Ac{fi!D mutant against those plant pathogens (Fig. 3).
[00111]
We further confin:ned that the APl 93At!,f'nD and
A~fjJ
mutants lacked symhesis
of difficidin by conducting LC>rv1S analysis of the cell-free TSB culture supernatants from
\.Vild-type APl93 and each of these mutants. A.s reported previously, only the deprotonatcd
fonn of oxydifficidin was detectable in hacterjal supernatants us.Ing 1vlS in the negative mode
([1vf --- HJ
=
559.3) (Chen et al., 2006}. with a molecular mass of 559.3 detected in
supenrntants of the \Vild-type AP 193 culture but not observed from Lhe culture of the Ae:(f!JD
mutant (Fig. 4) or from the A.~lp mutant (data not sho\vn). The 1\sr/AA mutant exhibited
difficidin syntht$iS as in the \Vild-type AP 193 culture (data not sh(nvn). These findings
dernonstrate the irnportance of difficidin in i.he hioco.ntro! acfrvity of subsp. pian!arun1 strains
against plant pathogens.
[00112]
axo1u.~mJdis
Strain AP! 93 secondarv metabolites control bacterial spot caused by A~
P'-l. vesicatoria in tomato plants. To determine the role of bioactive compounds
produced by strnin A.P 193 in providing protection against plant diseases, the AYJ 93 wild-type
strain and its AP l 93A((ff1D, AP! 93/\.tfiJ and AP l 93AszjAA mutants \Vere applied to tomato
plants several days before those plants \'Vere subsequently inoculated with plant pathogen .\::
axonopodis pv. vesicatoria< Both AP 193 \Vild-type and A.Pl 93As1i'4A significantly (P < 0.05)
reduced disease severity of bacterial spot on tomato plants compared to the disease control
(Table 3). Additionally, the application of strain APl 93 significantly increased the root dry
\.veight of the plants (Table 3). Unlike AP 193 \Viki-type and its APl 93As1/A.A mutant, strains
API93A~li) and AP1936c{/'1D neither protected tomato plants from severe bacterial spot
caused by X axonopodis pv, vesicatoria nor improved plant gn)\Vth (Table 3), further
supporting the importance of difficidin for plant disease protection. These findings are in
agreement \vith the in vilro antfoiosis pattern of APl 93 wild-type strain and its APl93/\({.fhD,
AP I 93A4j1 and /\P 19 3AwfAA mutants demonstrated against plant pathogen X axonopodis
pv. vesicatoria.
38
WO 2016/054222
PCT/US2015/053239
[00113]
Discussion
[00114]
PGPR Haci/lus spp. strains are used
\VOrldwidt~
to improve crop yields
~md
to
protect against plant diseases. In this Example,. 12 PGPR genomes were sequenced, including
B. subtilis, B. pumilus, B. arnyfoliquejcJCiens, B.
tJl(~javensis,
B. siarnensis, B. sonorensis, and
B. tequilensis. These data \Vere analyzed using ANI, gyr.B-hased phylogenies and core
genome-based phylogenies to resolve taxonomic affiliation of Bacillus spp. strains. Our
findings demonstrate that half of the strains sequenced in this Example are affihated \.vi th B.
amylofiquejdciens subsp. p!antarum, including strain (iB03 that vrns formerly designated as
B. subtilis. Previously, B. siamensis type strain KCTC 13613T \.vas proposed as a novel
species (Sumpavapo! et al., 2010), but a Bacillus core genome-based phy!ogenomic analysis
(Fig.
l)
n.~'Vt~aled
that B.
siamensis KCTC
l 3613T is instead affiliated v. ,ith B.
amJ•lofiq1u-'.fi.rciens subsp. plantarum. This finding supports the results of Jeong et al (Jeong et
aL 2012) that determined the close affiliation of B. siamensis type strain KCTC 13613T to B.
amyloliquefaciens subsp. p!antarum based on ANl. These findings also support the continued
use of core genome-based phylogenomic approaches to provide better phylogenetic resolution
than analyses that use a single housekeeping
g'-~n'-~
(e.g., gyrB). Phylogenies based on gyr8
and core genome sequences demonstrate that B. amyloliquef(.1ciens suhsp. plan!arwn are
highl'y similar, but i;.:omparison of their prowomes demonstrates that they are closely related,
yet distinct and may exert plant grmvth-prmnoting activities through different mechanisms.
[00115]
B. <w1..i·lo!iqulfin:iens subsp. piantarmn strnin AJ30 l \Vas isolated from t.he
intestine of channel catfish (Ran et aL, 2012), but its affiliation \Vith plant-associated strains
inay suggest transient presence v•iithin a fish gastrnintestinal tract; hmvever, given that the fish
feed is soy-based it is likely that the plant-based diet \Vas also a factor in the growth of this
strain \Vithin a fish intestine. Similarly, B. siamensis type strain KCTC l3613T was found to
he closely affiliated with B. amy!oliquqfaciens subsp. plantarum and \Vas isolated frorn salted
crab, rather than a plant-associated source. The efficm::y of strains A. BO I, API 93, and other
plant-associated strains as prohiotics in fish shows the capacity for biocontrol of animal and
plant pathogens as \veH as an overlap in host colonization (Ran et aL, 2012).
39
WO 2016/054222
[00116]
PCT/US2015/053239
\Vith rapid advances in sequencing technologies it is no\v possible to extend
genomic analysis beyond individual genomes to analyze core genomes (!vledini et al., 2008)-
ln this Example, core genomic aimlyses \Vere conducted on PGPR strains from species
affiliated v,..-ith the B. subtilis group. This analysis identified 73 genes exclusively present
among all subsp. plantarum that are absent in subsp. amyloliquejhciens strains. This small
munber of subsp. plantarum-specific genes agrees \Vith a previous report that identified 130
subsp. plantarum-specific genes using a limited number of genome sequences from subsp.
p!amarum strnins (He et al., 2012). Of tl1es(' 73 plamanmH;pecific genes identified in this
Example, many are predicted to he important for plant-associated and soil-associated
funct1ons. For example, genes that are required for the use of D-ga!acturonate and Dglucuronate \Vere found in the pool of 8- amy!oliquefaciens subsp. p!antarum-specific core
genes. This observation is consistent \:vith the absence of these genes in the genome of B.
amy!o!iq1u~f2r.ciens
subsp. am:vloliqtHfaciens DSlv17 (Ruckert et aL, 201 l ), a strain \Vithout any
reported PGPR activityJ>ectin, a complex polymer found in plant tissues, is broken dmvn to
D-i:z.lucmrmate and D-galacturonate \Vhich then serves as a carbon source for bacterial growth
.....
ot.,..
--
(Nemoz et aL, 1976). This pectin could potentially serve as a nutrient source for efficient root
colonization of PC!PR through competitive nutrient uptake. Therefore, the presence of genes
that enable
D-ga!acturonate and D-glucuronate utilization could be advantageous for H
1.unyloliq1.u:fc1ciens subsp. plamarwn for plant growt.h-prornoting activity through efficient
root colonization.
[00117]
Since 1nany of the PGPR strains are fron1 the B. suhtilis group, the core
genome estimation \Vas expanded to include a larger number of B. subtilis strains. Increasing
the number of Bacillus subtilis genon1es analyzed to 53 resulted in a 579,166 bp core genon1e
that is predicted to encode 674 ORFs. This smaller number of predicted genes reflects
genomic diversity among the B. subti!is group. This finding demonstrates that the number of
ORFs fr)tmd in the B. subtilis group core genome is dose to the number of B. subtilis ORFs
that are considered as :indispensable for growth in complex media (610 ORFs)
(htlp:/h'<-'V-i\V .n1inibacillus.org/proi(~ct#ge11es).
40
WO 2016/054222
[00118]
PCT/US2015/053239
To ·validate a gene's involvement in plant-related processes, h 1s essential to
construct isogenic mutants that are devoid of those genes, Therefore,
\Ve
deleted genes from
PGPR strain AP 193 to evaluate tl1e role of secondary 1netabolitc biosymhesis gene clusters in
the biologkal control of plant paihogens. To do this, a rnethy1ated shuttle vector pNZTl
(Zakataeva et aL, 20 l 0) with gene deletion constructs delivered targeted genetic modifications
to APl 93, demonstrating the effic.:acy of in vitro m.ethyhnion of plasmids by cell-free extract
in circumventing a restriction system that \Vas presumed to have prevented transformation
through electroporation.
[00119]
Difficidin is a highly unsaturated 22-rnembered macrocyfo.: polyene lactone
phosphate ester \Vith broad-spectrurn antibacterial activity (Zimmerman et aL, 1987).
Difficid.in e;9ressed by strain FZB42, together \.Vith the dipeptide baci1ysin, are mnagonistic
against
Fnvinia amvlovora --- the causative agent
of fire blight
disease in orchard trees (Chen
......
.......
......
~·
et aL, 2009), This Example using an isogenic rnutant APl 93l'1t:{fl1D demonstrated for the first
time that diliicjdin solely,, not in conjunction \vith Hny other polyketfrles or dlpeptides, exerts
in vitro antibacterial activity against plant pathogens, such as Pseudomonas SJTinge pv _
tabaci, Rhizobium radiobacter, ){amhomonas axonopodis pv _ vesicatoria and Xanthomonas
fft.:onopodis pv_ campestris. \Ve also demonstrated, hy using isogenic nwtant i\Pl 93/"~(tfh!.),,
that difficidin expression is responsible fbr control of bacterial spot disease in tomato plants
caused by .A~ .-o.:onopodis pv, vesicatoria. Taken together, these findings demonstrate that
diftkidin is the most important strain AP193 secondary metabolite for biological control of
plant djseases due to bacterial pathogens. In addition, the construction of the ,\/JJ gene deletion
alk)\ved investigation of multiple secondary metabolites
prnducz~d
hy AP 193 and their
individual contributions to biocontrol activity. The 4}J deletion mutant lost antagonistic
activity against each pathogen that \vas susceptible to the AP 193 \vi Id-type strain. f\.fotants
with the 4j:i deletion are expected to lose the ability to synthesize difficidin in addition to
other metabolites. Because the lack of anthnicrobial activitv of AP193A~f.b is consistent \Yith
~·
.. J A
that of the APl 93/\l{fiiD mutant, this therefore suggests that difficidin is the prirnar.y
metabolite responsible for in vitro inhibition of bacterial paihogens. In contrast, the surfactin
mutant retained antimicrobial activity against an plant pathogens tested, demonstrating that
41
WO 2016/054222
PCT/US2015/053239
surfactin is neither cdtical for in vitro antib1otic activity nor
1ntlu~mces
the synthesis or
secretion of other secondary metabolite biosynthesis in this Bacillus spp. strain; ho\vever,
surfric.tin may influence plant gn.i\vth promoting activity since it has been observed that
smiactin of B. subtilis elicits JSR in plants (Ongena et aL, 2007) and is expressed in the plant
cells colonized by FZB42 (Fan et aL, 2011 ).
[00120]
By studying the contributions of genetic 1oci that are conserved among top-
perfonning PGPR strains
Vile
continue to uncover the relative contributions of genes in plant
colonization,. growth promotion, and/or pathogen hiocontroL In particular, future investigation
of genes related to the uptake and use of pectin-derived sugars \vill hdp determine the rdative
importance of these genes for colonization of plants and persistence within this microhiome.
Comparative genomic analysis of Bacillus spp. PGPR strains has led to a better understanding
of gene products and provides a foundation to develop application strategies that result in
greater plant growth promotion and biocontrol activity.
[00121]
Tables
[00122]
Table l. Bacterial strains and plasmids used in this Example.
42
WO 2016/054222
PCT/US2015/053239
······················ Rekrnnt charncteristi~'<;
S()(ir(:(;·(;i:i:(:!c~1·ei1c(:······················]
E coh Kl 2 ER2925
dcm-6 dmu l 3 .. Jil9
N("»,· Eiwbnd B1olabs
,•..................................................................
;~ .................................................................
:•. I51~:·'};:;3~;;r;·I<fo;;r;1;~;
.......................1!
[)t~«~il!~.~ii:.pl;1~1~1i~i~
i H a.>n).foliq.w.fociens sub:->p
· plamarnm strain APl93
: {Department of rintomolugy
• and Plaut Pathok1>rv. Auburn
Wild type
I
!
J
T::!1~i_\'~:r_s_il_yt............~:~......................
• (k:ficknt in li1w1K'.t)ti(k-:;
•: . . 11. · c;tu·'v
,
· .t
:ii
• ·:111·! 1·1lvk·,.,;;,l··c;
: 11· "" ''..
:------------------------------------------------------------------~--~---:~... L~_~:....~----~2:.:~:~::............................ ~---------------------------------------------------------------------~
·
~ defici<.:ni !n :->urfadin
•
!i
APl93/1.s.>f4A
:
d (
. Thie; siudy
.................................................................._l ...................................................................
• \. , .)., \ .;;'n
..
1 ·1· ··· :h
: ·
:
"
-
1
)..'
:
................................................................ i. pr'} •. t1~.1l:>t1..
• APJ'} 3 \,.LliiD
..................................... '. ............................................
· deficient in diffiddin
•This '>tudy
,............................................................. p1ycl1tt;~i1:11~ ..................................... ...............................................
i l?acillu.~· an1yloli1..JU(;.j(icicns :
i (·t-~· ., .:.,;~ .;,.)· ( ·1 )i}llll,"
1 type
• FZB42
' \\"id
• .._, ..1:,1 ' " ,1 ., _,' . ,,,.
1
!
I
i
j
I
:--~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~:-~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~-l;
·
: .E'. cofi~1J~:u.~illus shuttle
·
p~lK1
! plasmid, rolling ('.irde
BGSC
..................................................................1ypli~~1iiy~,.91~r~ ..
~ Rqllkution
! th<.:nno,,;ensiiiv<.: deri,·ntive
pNZTl
d' ihi;O' wlling-circk
pla:->nud pV/VOl (po·•·
t
Xi<w:dwu Zkmg, Vu:gmia
Tech
rnlii:.~~1n
.................................................................,.....
l ....... :....c..FrnH)
::........ :............... .
~ pNZ.T 1 wi ih up::>trcam and
•This studv
..................................................................L.~~~h~·ii:'~~Tiid~;~~;~.-i~~~;;~·;··················································································I
• pNZ~s.rf
pNZ~dif
1constrnd of sr.fAA
This. study
···········································r·!)i\;'jji. \~:j{j1.k;};)~{~~)tii
• ·11~i~·~~i1~l;:··········································1
.................................................................~ ~(~l1S:t1:1~~1.(~i'..(tri1P ............................................................................................
43
'
!
!
WO 2016/054222
[00123]
PCT/US2015/053239
Table 2. Summary of draft geno1nes of Bacillus specfos sequenced nsed in this
Example
·l=l~t., . ,.
:!:: . ~;~;::t~l···, ~:· 1·~~.~~:.::::i:;,;~.··1······~i~~i~;;;:~:········· ~1:~~···_1_.N_:_,_,_:_,1_,_:_;_;,_~_l_.~_.,_:_,_'._:_1_._._!',',
'•(i
(>{kb)
asstmbly.)
.
Accession No.
ABIJ1
20
3303 . 2l.16
46.4
A.P79
47
,_
45.8
...i~ ..
..{\!'7..L... ....J?.L...... __4.;, _~;.~-.:_·,'.'.(~_'):-'.l:~t_ .:~j:..+ .. 4 s 7 ::T:
.
~~:~: ~~;
.
:
~-~
----·+·
.
,
~:~;~):~~~:
.. , ...
.PRJNA2393 l7
SRX475739
pRJNA23 9) l7 ..., ......\f<![~?~.Z~_()_______..
PRJNA.2:'9317
SRX475741
~/)
',._>T{.!· .·,.:·'
44
3944
.........!?............. :::::4}{C:)_
1;;} .·:·i~*::·~-~~:;:~·;~"{f; . .,. . . ;;~i~;;~;~; .
:~I
;~
4J6i'i
. . . }{;}······1
'~~~~\'.F??~{~~:~~~------5<)""""""""''""':1}i:~;DfFY""""C""'TGf""'''~~~Ei~JB'.S?:~v~)T~~~:·~~~~~~~Fgay~};(j[~~~T~~~~~~~~~?V~~~~~~~T~~~~~~;1yrr~~~~1
GB03
26
:t849547
46 -s
PRIN\!17787
"Rxu:nrnn
"60
392!': :
~'''''''''''''''''''''~
~
~''''''''~''''''''''''~,,,,,,,,,-,~.":,,,,,,~,,,,,,,,,,,~,.~,,,~,,,.:7~::.~.,-:,,,,~,,.,,~,,,,,,,,::,,,,:,,,~~.::.~.~::::.-~,,,,,,,,~,,,,,,,,,:,,-~,,,,,,,,,,,,~,,,,,,,,,,,,,,,,,,,,,,,,,:
.••••••••••••••••••••••• •••••••
. INR7
~~~!~~:
.
44
. }/-l~l)!l?
l;
.
~;335.582
~
41 :;
.·.·.·_4_:_·_,_>_._•.
. PRJN:\22778() .
SRX447924
.
750
.
3857
....................................... ·~ ....................................... ·:· ....................... ~·
PRJNA16!489
SRX450083
500
3915
J·FiNA227 791 ;
4299
j
~.!_···················J~J·!.T~,,~;;;;.~~.i····! ·······~-~~~~;));~.;········ ·······~~;~·········· · · · · · · 4· ·.·_J_._4_· · ·(· •.~.·.·.·.·.·.·.1, ,
'· .................... ·:· ......................t...... :..................... ~
K.crc
n
3.779,696
46.:>
:. . ~~:;:1.r--l·-·······.~-~---······j····:~-,:~-;~,;~~-;····: . . ._;·;··~---·i . . i~i~~;~~-;~;-~·;;·~~----i·-······~-~~~~;~~~~-;~:;·······:········;·~~;~~~~---····j·······~;~~;~;·······I
I
~~i~I
;":
1981302 ; 43 ')
44
SRX450086
"
l ()OlJ
"
I
WO 2016/054222
[00124]
PCT/US2015/053239
Table 3. Effects of plant
gro\vth~promoting
rhizobacteria (PGPR} strains on
severity of bacterial spot disease and plant growth
Disea;,;e St'.verity '
Shoot Dr;./ \Veight {g)
Root Dry \Vcighl (g)
Disease Control
2.1 l a
L07 he
0.378 c
AP19J
1.30 b
2.18 b
OASJ
AP.l 931ls1jAA
1.48 b
2.16 b
0.423 abc
/I.Pl 93l':>..~fj)
2.31 a
2.18 b
0.405 ahc
AP 193/ld{l
2.06 a
2.00 c
0389 be
O.OOc
2.38 a
OA-35 ah
0-35
0.15
0.050
Ht.~althy
Control
LSD
H
Note:
a. The experimental design \Vas n randomized cumpkte blnck \Vith ten n:p!icatiuns per
treatmenL The CXJKTinient ·was conducted twice. Values fr.1lk)\\'t'.d by the smm'. ktkr
·were not significantly drffi::r.:.'.nt (Fb0Jl5) according to F1sd1er's p1uleckd LSD.
b One plant 1vas in t~ach replication. P!nnts \\·1.:~re sprayed with PGPR susrKnsion (106
Cfl)/ml) one 1vcek atkr transplanting, and \Vere challenge-inoculated 'Nith pathogen
solutions (1 o' CFUhn!) three days afk'.r inoculating PGPR.
c. Di;;,ease severitv ratimis and lrnn.'t:'.St 'Ntre done 14 davs later. For di;:,ease stveritv
rating, fbur compound leafs \Vere sdcct~xl from the bottom of each plant. The disease
S('.\'Crit:y of each of t!K~ compound ka.,..-es \Vas ddennincd by rating HK~ disease S(.'vcrity
nf each kat1et and cakulating the avcn1ge rating for tht:'. tompound leaf The leaflet
\Vas rated using a 0-4 rating scak, ,,·here 0 heaHhy kafleL l <20':\~) necrotic area of
the leaflet, 2 '" 20-50'Vi; necrotic area of the leaflet, 3'" 51-80 l\{, necrotic area of the
k'.afkt.. 4 "' 80-100 '\, nccroli''. area of the leaflet, or fiJHy dead !eafid.
~
~·
~
y
000
00'
[00125]
References
[00126]
Alegre, MT., Rodriguez, MC., and ~,.fosas, J)vl (2004). Transfbrmation of
Lactobac.i!Jus plantarum by electroporation with in vitro modified plasmid DN;'\. FEMS
idicrobiol Leff 241, 73-77. doi: 1O"1016.. j.fernsle.2004. l 0.006.
45
WO 2016/054222
[00127]
PCT/US2015/053239
Aziz, R.K., Bartels, D., Best, A.A., Dejongh, M., Disz, T., Ethvards, R.A.,
Formsma, K., Gerdes, S., Glass, E.M., Kubal, rvl., l\·foyer, F., Olsen, G.J., Olson, R.,
Ostem1an, A.L., Overbeek, R.A., !vlcneil, L.K., Paarrnann, D., Paczian, T., Parrello, R,
P11sch, G.D.,, ReidL, C., Stevens, R., Vassieva, 0., Vonstein, V., \Vilke, A,, and Zagnitko, 0.
(2008). The RAST server: Rapid annotations using subsystems technology. BA!C Genomics
9:75. doi: Artn 75 Doi 10.1186./1471 -2164-9-75.
[00128]
Bhn, K., .~·fedema, l\lH., Kaze1npour, D., Fischbach, l'Vl.A., Breit!.ing, R.,
Taka1H.\ L, and Weber, T. (2013). antiSl\.'1ASH 2.0······a versatile platforrn for genome mining
of
secondary
metabolite
producers.
/Vudeic
Acid'>
Res
41,
W204- \\/212.
doi:
10.1093/nar/gkt449.
[00129]
Borriss, R, Chen,_ X.H., Rueckert, C., Blom, J., Becker, /\., Baumgarth, R,
Fan, B., Pukall, R, Schumann, P., Sproer, C., Junge, H., \later, J., Pnhlec. A., and Klenk, H.P.
(2011 }. Relationship of Bacillus amy1oliquefaciens clades associated '>-Vith strains DS!vt 7T
and FZB42T: a proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov.
and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome
sequence compadsons . .lnr J .\vst Evol Microbiol 61, 1786-1801. doi: 10.1099/ljs.0.023267-0.
[00130]
Chen, X.H., Kournoutsi, A., Scholz, R, Eisenreich, A., Schneider, K.,
Heinemeyer, l.,
~Jorgenstern,
B., Voss, B., Hess, \V.R., Reva, 0., Junge, ff, Voigt, B.,
Jungb!ut, P.R., Valer, J., Sussmuth, R., Liesegang, ff, Strittmatter, A., Gottschalk, G., and
Borriss, K (2007a) . Comparative analysis of the (.:omplete genome sequence of the plant
growth-promoting bacterium Bacillus mnyloliquefaciens FZB42. Nat Biotechnol 25, 10071014. doi: l0.1038/nht1325.
[00131]
Chen. X.H., Koumoutsi, A., Scholz, R., Eisenreich, A., Sch.ne.ider, K.,
Heinemeyer, L, .l'vlorgenstern, B., Voss, R, Hess, vV.R., Reva, 0., Junge, H., Voigt, B.,
Jungblut, P.R., Vater, .L, Sussmuth, R.,. Liesegang, H., Strittmatter, /\., Gottschalk, G., and
Boniss, R. (2007b} Compa.rati"\'e analysis of the complete genome sequence of the phmt
46
WO 2016/054222
PCT/US2015/053239
grc.n.vth-promoting bacterium Bacillus arny!oliq1.{(fbcie11s FZB42. Nat Bioteclmol.
25~
l 007-
1014. doi: I0.1038/nbtl325.
[00132]
Borriss, R.
Chen, X.l-L Scholz,
re
Borriss, 1vL Junge, H., l\dogeL. G., Kunz, S., and
(2009). Difficidin and bacilysin produced by plant-associated Bacillus
arnyloliquefaciens are efficient in conlTolling fire blight disease. JBiotedmol 1.40, 38-44. doi:
10.1O16/j jbiotec2008. 10.015.
[00133]
Chen, X.H., \later, J., Piel, J., Franke, P., Scholz, R., Schneider, K.,
Koumoutsi, A., Hitzeroth, G., Grnmmel, N., Strittmatter, A.\V., Gottschalk, G., Sussmuth,
R.D., and Borriss, R. (2006). Structural and functional characterization of three polyketide
synthase gene dusters in Bacillus amyloliquefociem; FZB 42. J Bacterial l 88, 4024-4036.
doi: 1O.l128/JB.00052-06.
[00134]
Chin, A., F., \V .T., Bloemberg, G.V., tvlulders, I.I-I., Dekkers, L.C., and
Lugtenberg, BJ (2000). Root colonization by phenazine-1-carboxamide-producing bacterium
Pseudornonas chlororaphis PCLl 391 is essential for biocontwl of tomato foot and root rot
klol Plant Aficrohe lnleract 13, 1340-1345.
[00135]
Choi, S.K., Jeong, El, Kloepper, J.\V., and Ryu, CJvL (2014). Genome
Sequence of Bacillus amyloliquefacicns GB03, an Active lngred1ent of the First Comrnen.:1a1
Biological Control Product. Genmne Announc 2, 01092-01014.
[00136]
Darling, AC., 1'vfau, B., Blattner, F.R., and Perna,. N.T (2004). Mauve:
multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14,
1394-.1403. doi: 10.1101 /gr.2289704.
[00137]
'k-.ers, r,. C'"' "'1
·1 r:.,
r1 l)l··
i· 1
(.,
·C' ., C't.
I)eK
iv uId ers, ..
..)Oeiic.11,
.....
. 11m,·
~ ·\'Y.
J 'I' ,,.,
r· .,
h.,
1
\-i,.,.,~rtJes,
.. "
~ 1·:1,,
!-\,
1
and Lugtenberg, B.J. (2000). The sss colonization gene of the tomato-Fusarium oxysporum f.
sp. radicis-lycopcrsici biocontrol strain Pseudomonas fluorescens \\."CS365 can improve root
colonization of other 'wild-type pseudonmnas spp.bacteria. :\.fol Plam A:ficrobe !merau 13,
1177-1183. doi: 10.1094/f\·1Pl\.1 l.2000.13. I I. l 177.
47
WO 2016/054222
[00138]
PCT/US2015/053239
Dietel, K., Beator, B., Budihmjo, A., Fan, B., and Borriss, R. (2013). Bactedal
Traits Involved in Colonization of Arah1dopsis thaliana Roots by Bacillus amylolique;_Hwiens
FZB42. Plant Palfwl J 29. 59-66.
[00139]
Emmett, E.a.B., Klimo\vicz,. A.K., Thomas, !'vLG., and Handelsman, J. (2004).
Genetics of bvitterrnicin A Production by Bacillus cereus. Appl Environ Microbiol 70, 104113. doi: l0.1128.iaem.70.Ll04-l 13.2004.
[00140]
Fan, B., Chen, X.H., Budihai:jo, A., Bleiss, W., Vater, J., and Borriss, R.
(2011 ). Efficient colonization of plant roots by the plant gn.nvth promoting bacterium Bacillus
amylo!iquefadens FZB42, engineered to express green fluorescent protein. J Biolechnol 151,
303-31Ldoi:10.10Hi~j,jbiotec.2010J2.022.
[00141]
Felsenstein, J. (1981 ). Evolutionary trees from
DNA~
sequences: a maximum
likelihood approach. J kfof Evof] 7. 368-376.
[00142]
Friis, C., \Vassenaar, T.l\:1., Javed, !VI.A., Snipen, L, Lagesen, K, Hallin, P.F.,
Ne\velL D.CL Toszeghy, M . , Ridley, A, lVlanning, G., and Ussery, D.W. (20to). Genomic
characterization of Campylohacter jl;/uni
strain
~H.
PLoS One 5, el2253.
doi:
10.137LJournaLpone.0012253.
[00143]
Goris,..L, Konstantinidis,. KT., K!appenbach, J.A., Coenye, T., Vandamme, P.,.
and Tiedjt\ JJ\L (2007). DNA-DNA hybridization values and their relationship to w·holegenorne sequence s.irnilarities. Inf J S)wt Evol Alicrobiol 57, 81 ¥9 l.
[00144]
Hao, K., He, P., Blom, l, Rueckert, C., Ivlao, Z., Wu, Y., He, Y., and Borriss,
R. (2012). The genome of plant grov-ith-prom.oting Bacil1us amylo1iquefaciens subsp.
p/antarum strain YAU 89601-\'2 contains a gene cluster for rnersacidin synthesis. J
Bacteriol 194, 3264-3265. doi: HU 128/JB.00545¥ 12.
[00145]
He, P., Hao, K., Blom,. l, Ruckert, C., Vater, .I., l\,lao, L, \\iu, Y., Hou, rv:L.
He, P., He, Y., and Borriss, R. (2012). Genome sequence of the plant grmvth promoting strain
48
WO 2016/054222
Bacillus
PCT/US2015/053239
amyloliqud~-tciens
subsp. p!antarum B960 l -Y2 and expression of 1nersac1d1n and
other secondary metabolites . ./ Biotechnol 164, 281-29 l.
[00146]
Horton, RJvL, Hunt, H.D., Ho,, S.N., Pullen,, J.K., and Pease, L"R. (1989).
Engineering hybrid genes \:Vithout the use of restriction enzymes: gene splicing by overlap
extension. Gene 77, 61-68.
[00147]
Hossain, r...tJ., Waldbieser, G.C., Sun, D., Capps, NJ(,, Hernstreet, \:"\/J3.,
Carlisle, K., Griffin, MJ., Khoo, L., Goodvtin, A.E., Sonstegard, T-5., Schroeder, S., Hayden,
K., Nevdon, lC, Terhune, lS., and Liles, M.R. (2013). Impllcation oflaternl genetic trnnsfor
in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish.
PLoS One 8. e80943. doi: I 0.137 Ljournal.pone.0080943.
Idriss, RL, Makare\vicz, 0., Farouk, A" Rosm.~r, K. , Greiner, R., Bochtnv, IT,
[00148]
Richter,
T.,
and
Boniss,
R.
(2002).
Extracellular
phylase
activity
of
BaciHus
arnyloliquefaciens FZB45 contributes to its plant-gr0'-\·1h-promoting effect. Aficrobiology 148.
2097-2109.
[00149]
Jeong, H., Choi, S.K., Kloepper, J.\V., and Ryu, CJvL (2014).
c;,~norne
Sequence of the Plant Endophyte Bacillus pum.llus INR7, Triggering Induced Systemic
Resistance :in Fidd Crops. Genome Amwunc 2, 01093-01014.
[00150]
Jeong, H., Jeong, D.-E., Kim, S.H., Song, G.C, P<.uk, S.-Y., Ryu, C-!VL, Park,
S.-1-I., and Choi, S,-K, (2012). Draft Genome Sequence of the Plant Growth-Promoting
Bacterium Bacillus
siamensis
KCTC
l3613T.
J Bacteriol 194, 4148-4149. doi:
10.1128.:jb.00805-12.
[00151]
Ka1nilova, F., \lahdov, S., Azarova; T.,
~folders,
l.; and Lugtenherg, B.
(2005). Enrichment for enhanced cornpetitive plant root tip colonizers selects for a nev·.i class
of biocontro1 bacteria. Environ lvflcrohiol 7, 1809-18 l 7,
49
WO 2016/054222
[00152]
PCT/US2015/053239
Kim, RK., Chung, J.H., Kim, s.·Y., Jeong, H., Kang, S.G., K\von, S.K., Lee,
C.H., Song, J.)''., Yu, D.S., Ryu,_ C.M., and Kim, JJ·'. (2012). Genome sequence of the leafcolonizing Bacterimr1 Bacillus sp. strain 586, isolated frorn a cherry tree. J Bacteriol l 94,
3758-3759. doi: 10.1128/JB.00682-12.
[00153]
Kloepper, J.\V., Ryu, CJVL, and Zhang, S. (2004). Induced System.ic
Resistance and Promotion of Plant GrO\vth by Bm::illus spp. Phytopatho!ogJ' 94, 1259-1266.
doi: 10.1094/P.ITYT0.2004.94. l L 1259.
[00154]
Kloepper,
l\"\/.,
and
Schroth,
M.N.
p978).
Plant
growth-promoting
rhizobacteria on radishes. Proceedings 1f the 4th International Co1?f'erence on Plant
Pathogenic Baeleria. Station de Patho!ogie Vdgha!e et Phytobacth·iologie, JARA, Anger».
France
2~
879---882.
[00155]
Kokalis~BureHe,
N., Vavrina, CS.,
Rosskopf~
E.N., and Shelby, R.A. (2002).
Field evaluation of plant gro\:vth-promoting Rhizobm:.:teria amended transplant mixes and soil
solarization for tomato and pepper production in Florida. Plant and Soil 238, 257-266.
[00156]
Letunic, L, and Bork, P. (20 l 1). Interactive Tree Of Life v2: on line annotation
and display of phylogenetic trees made easy. Nucleic Acids Res 39, \.\/475-\\/478. doi:
10.1093/nan'gkr201.
[00157]
Lugtenberg,
Rhizobacteria.
Anmf
B.,
and
Rev
Kamilova,
F.
Microhiol
(2009).
63,
Plant-Growth- Promoting
541-556.
doi:
doi: !O. ! !46.iannurev.micro.62.081307. !62918.
[00158]
for
gent~
[00159]
Lukashin, A.V., and Borodovsky, !'vL (1998). GeneMark.hmm: nev.' solutions
finding. A'ucleic Acids Res
26~
1107-"I 1!5.
J\,fodini, D., Sermto, D., Parkhill, J., Reiman, D.A., Donati, C, \Joxon, R.,
Falk.ow, S., and Rappuo!i, R. (2008). Microbiology in the post-genomic era. 1Vai Rev
kficrobiol 6, 419-430. cloi: l OJ 038/nrmicrol 901.
50
WO 2016/054222
[00160]
PCT/US2015/053239
Nakkeeran,. S,, Fernando, \V.GD., and Siddiqui, LA. (2005). "Plant gro\\·th
promoting rhizobacteria formulations and its scope in commercialization for the management
of pt$tS and diseases," in PGPR: Biocontro! and Biqj(:rtilization, ed. Z.A. Siddiqui.
(Dordrecht, The Netherlands: Springert 257-296.
[00161]
Nemoz, G., Robert-Baudouy, l, and Stoeber, F. (l 976). Physio!og1cal and
genetic reb:rtllation of the aldohexuronate transport system in Escherichia coli. J Bacterio/ 127,
706-718.
[00162]
Ongena, :tvt, Jourdan, E., Adam, A, Paquot, ~vL, Brans, A., Joris, B., Atpigny,
J,L., and Thonart, P. (2007). Surfactin and fengycin lipopeptides of Bacillus subtl!is as
elicitors of induced systemic resistance in p!auts. Environ Microbiol 9, 1084-1090. doi:
l 0.111 lij .1462-2920.2006.01202Jc
[00163]
Pfeiffer, W., and Stm.natakis, A. (2010). "Hybrid MPl/Pthreads Parallelization
of the RAxfv1L Phylogenetics Code. In: Ninth IEEE International Workshop on High
Perfonnance Con1putational Blo!ogy
[00164]
Podile,
A.R.,
and
(HiC0~·1B
Kishon.\
2010}" (Atlanta, USA).
G.K.
t2006).
"Plant
growth-promoting
rhizobacteria," in Phmt-_Associated Bacteria. ed. S-5. Gnanamanickam.
(Netherlands:
Springer), pp. 195-230.
[00165]
Ran, C, CatTias, A, \Villiams, !VLA., Capps, N., Dan, B.C, Newton, J.C.,
Kloepper, J.\V ., Ooi, E.L., Brmvdy, C.L., Terhune, .LS., and Liles, 1\.-LR. (2012). Idenlificatfrm
of Bacillus strains for biological control of catfish pathogens. PLoS One 7, e45793. doi:
l0.137lijoumal.pone.0045793.
[00166]
Reva, O.N., Dixdius, C., TV!eijcr, J., and Priesc F.G. (2004), Taxouomic
characterization and plant colonizing abilities of some bacteria related to Bacillus
amyloliquefaciens and Bacillus subtilis.
FEAlS Microbiol Ecol 48, 249-259. doi:
I 0.1016zi .femsec.2004.02.003.
51
WO 2016/054222
[00167]
PCT/US2015/053239
Rkhter, J\t, and Rossell6-M6ra, R. (2009). Shifting the genomic gold standard
for the prokaryotic species definition. Proc l1latl Acad Sci [l S A l 06~ 19126-19131. doi:
l 0.1073/pnas.0906412106.
[00168]
Ruckert, C, Blom, J., Chen,. X, Reva, 0., and Borriss, R. (2011). Genome
sequence of B. amyloliquefaciens type strain DS:Vl7(T) revea!.s differences to plant-associated
B. amylohquefaciens FZB42 . .J Biotechnol 155, 78-85. doi: 10. IO 16/j ,jbiotec.2011.01.006.
[00169]
Ryu, C-i\.L,. Farag, !VI.A, Hu, C.-H., Reddy, ivLS., Kloepper, J.\V., and Pare,
P.\\.t. (2004). Bacterial Volatiles frlduce Systemic Resistance in Arabidopsis. Plant Physiol
134; I017-l026. doi: 10J 104.ipp.103.026583.
[00170]
Saitou, k,. and Nei, M. (l 987). The neighbor-joining method.: a new· method
for reconstructing phylogenetic trees. A.fol Biol Evo 4, 406-425.
[00171]
Sumpavapol, P., Tongyonk, L, Tanasupmvat, S., Chokesajjawatee, N.,
Luxa.nanil, P., and Visessanguan, \V. (2010). Bacillus si.amensis sp. nov., isolated frorn salted
crab
(poo-khemi
in
Thailand.
int J 5)wt
Evol J.ficrobiol 60, 2364-2370.. doi:
10.1099/ijs.0.018879-0.
[00172]
Tamura, K, Peterson, D., Peterson, N., Stecher, G., Nei, !vi., and Kum.ar, S.
genet1cs analvsis using..,... n1aximum Likelihood.
{2011).
. !VIEGAS: molecular evolutionarv
....
......
~·
~
evolutionary distance,. and maximum parsimony methods. Mo! Biol Evo 28, 2731-2739.
[00173]
Xu, Z., Zhang, R., \Vang, D., Qiu,
~vL,
Feng, H., Zhang, K, and Shen, Q.
(2014). Enhanced control of cucumber \Vilt disease by Bacillus amyloliquefaciens SQR9 by
altering the regulation of Its DegU phosphorylation. Appl l·:'!1viron Microbio! 80, 2941-2950.
doi: lO. I 128.iAETVL03943-l3.
[00174]
Yuan, l, Raza, \V., Shen, Q., and Huang, Q. (2012). Antifongal Activity of
Bacillus arn_vloliqut;laciens NJN-6 Volatile Compounds against Fusarium oxysporum f. sp,
cubense, Appl Environ },,ficrobiol 78. 5942-5944. doi: 10, 1128.iaem.01357-12.
52
WO 2016/054222
[00175]
PCT/US2015/053239
Zakataeva, N.P., Nikitina, OS., Gronskiy, S.V., Ron1anenkov, D.Y, and
Livshits, V .A (20 l 0). A simple method to introduce marker-free genetic modHkations .into
the chromoson1e of naturally nontrnnsfi:m:nable 8acilius amyloliqrwfcu.:iens strains. Appl
Aficrobio! Bioleclmol 85, 1201-1209_ doi: 10.l007is00253-009-2276-l.
[00176]
(2011 ).
Zhang, G.-Q., Bao, P., Zhang,'\:'., Deng, A.. -H., Chen, N., and \\/en, T.-Y.
Enhancing
electro-transformation
competency
of
recalcitrant
Bacillus
amyloliqu4aciens by con1bining cell-\val! weaken.ing and cell-rnernbrane fluidity disturbing.
Anal Biochem 409, 130-l37. doi: http:.iidx.doi.on.v"l O. IO 16../Lab.2010.10.013.
[00177]
Zimmerman, S.B., Sch\vartz, CD., rvlonaghan, R.L, Pelak, BJ\., \Veissberger,
B., Gilfillan, E.C, l\fochaks, S., Hernandez, S., Currie, SJ\., Tejera, E., and Et AL (1987).
Difficidin and oxyd.ifficidin: novel broad spectmm antibacterial antibiotics produced by
Bacillus subtilis. L Production, taxonomy and antibacterial activity. J Amibiot (Tok_vo) 40.
1677-1681.
[00178]
Example 2 ~ Grant Proposal for "Pectin Stimulation of Plant Gr(nvth
('k
Disease Control bv Plant Grm.vth-Pronmting Rhizohacteria" submitted to i\labama
Agricultural Experiment Station
[00179]
Abstract
[00180]
P!a11t gro\vth-promoting rhizobacteria (PGPR) have been identified that control
plant diseases and pn.nnote O"\i·erall plant grow·th. "Rhizohacteria" means mot-colonizing
bacteria, and hence, root colonization is essential for pla11t grov~·th promotion by PGPR
strains. Plant roots
exmll~
various organic compounds, including sugars, and successful
bacterial colonization hinges on nutrient uptake from the host plants through extracellular
enzymatic activity. Strains of Bacillus amyloliqu(fhciens subsp. plan/arum (Bap) colonize
plant roots, and have been used as biofertilizers or biocontro! agents during the past decades.
Some of the best-perfom1ing PGPR Bop strains at Auburn have been subjected to a
53
WO 2016/054222
PCT/US2015/053239
comparntive genomic analysis, \Vhich indicates that
tht~
use of pectin is a conserved trnit
arnong these sequenced strains. As a stmctural component of the plant cell \Vall, much is
knovm regarding pectin bkKhemistry a11d plant synthesis; hO\vever, !itt!e is kno\vn about the
possible role of pectin in root colonization. In fact, the current scientific paradigm regards
pectin utilization as a function expressed by plant pathogens, and not as a potentially useful
characteristic expressed by plant-associated PGPR strains. \Ve no\.\,. have experimental
evidence that our best-perfom1ing PGPR Bap strains can obtain carbon and energy via 1)
productio11 of an extracellular pectinolytic enzyme(s) that degrades plant pectin into
hexuronate sugars, 2) transpo1t of pectin-derived sugars, and 3) utilization of these pectinderived sugars for bacterial respiration_ While these PCIPR Rap strains consistently perfrKm
,.vell under lab or greenhouse conditions, field trials are more variable in PGPR efficacy, \Ve
hypothesize that suppkrnenting pectin levels on
s(~eds
or in the plant rhizosphere \Vil! irnprove
the efficacy of PGPR strains in stim.ulating plant growth and disease control. Our specific
objectives to test this hypothesis are: i) Screen a large col1ection of Bap strains for pectin
degradation and utilization as a sole C source, 1i) Conduct a cotnparative genomic analysis
that includes Bap strains that lack the capacity for pectin utilization (see Example l above),
and iii) Evaluate pectin supplementation along ·with Bap stJains (\vith and without the ability
to use pectin) for soybean growth prornotion and disease cont:roL The informatfrm derived
from this study will provide, for the first time, an understanding of the growth-promoting
abilities of PGPR strains as int1uenced by pectin availability. The results of these experiments
'vil! provide the needed preliminary data in order
to
submit competitive proposals for
extranmrnl funding to federal agencies, particularly the USDA and the NSF, in which we
could extend these results to field settings \\-'ith multiple crop species. The practical use of this
information could enable sustainable solutions for biocontrol of agricultural pathogens and in
promoting plant gHnvth.
[00181]
Introduction
[00182]
There is a grO\vmg need for environmentally sustainable and effective
alternatives to antibiotic use in agriculture, The US Food and Drug Ad1ninistrntion has been
pursuing policies to reduce antibiotic usage in livestock, poultry· and crops ( l), and these
54
WO 2016/054222
PCT/US2015/053239
policies are expected to become more stringent in the US and other countries, This has
prompted the search fbr sustainable methods, like the probiotic strategies described in this
proposal, for the control of agricultural pathogens and enhancement of animal and plant
grmvth 'l.Vithout relying on antihiotics. Plant grmvth-promoting rhizobacteria (PGPR) have
proven to be promising as probiotic agents for the purposes of disease biocontrnl and
biofertilizers (2-5). While many specjes of hacieria are classified as PGPR strains, Bacillus
species have been closely studied due to their spore-frmning activity that confers a longer
shelf life <.md greater viability in comm.ercial biocontrol formulations. \Vithin thjs genus,
Bacillus anzyloliquejdciens subsp. plantarum (Bap) has recently emerged as an especially
effective PGPR spet:ies that lack any potential fbr pathogenesis (6, 7). At Auburn Universit.y,
Prof Kloepper has collected a large number (> 300) of PGPR isolates that show hiocontro!
and.ior plant growth-prornoting activities, of \vhkh 59 have been identified as Hap strains_
Working collaboratively \vith Prof. Liles, specific PGPR Bap strains have been identified
(thanks to previous A.AES and NSF support) to have the capacity to control disease in
channel catfish (8) and are being used in controlled trials in catfish-production ponds this
summer. Therefc1re, there is interest in using PGPR strains as alternatives to antibiotics in
agriculturally-relevant crops and animals, thereby increasing their effectiveness in these
agriculture and aquaculture applications.
[00183]
PGPR strains are considered biostimulants, in that they are microbial
inocula11ts that have beneficial probiotic interactions \Vith their plant host (9), For example,
inicrobial inoculants can solubilize phosphorus aml/or fix nitrogen that can then he absorbed
by plant roots, directly stimulating pla11.t growth. There is a large literature on the use of
bacterial inoculants for nitrogen fixation (10), and many Bacillus spp. strains have been
identified as phosphate-solubil.izing bacteria \Vith commercial potential as hiofertilizers (11),
In addition, PGPR strains have been fi:.mnd to produce many secondary metabolites that have
antibiotic activity against bacterial and/or fungal pathogens
O2-14 ),
and can also induce
control plant disease through production of compounds that induce plant systernic acquired
resistance (SAR; rnediated by salicylic acid) and induced systemic resistance (!SR; jasmonic
acid-dependent) mechanisms ( l 5-17), Tn fact, the collection of sequenced PGPR Bap strains
55
WO 2016/054222
PCT/US2015/053239
from the Kloepper laboratory includes strains that
havt~
been found to induce both plant
growth-promotion and disease control.
[00184]
The earliest reported studies of seed bacterization for agricultural purposes
dates to the use of Rhi::ohium inoculants on lebrumes in 1895 (18), Yield increases for cereal
crops afrer bacterial inoculants were applied 'Were observed in a vari.ety· of Soviet and Indian
studies throughout the 1960s a11d early 1970s ( 18). Hc.)\vever, field studies consistently
produced lo\ver yields than greenhouse studies, suggesting that the introduced microbial
population declined rapidly after soil inoculation (l 8). This decline \vas likely due to an
inability of the PGPR strnins to compete \Vi th the nonna! rhizosphere microbiota. Despite the
inherent difficulties of using bacterial inoculants, the biostimulant market in North America is
estimated to
annually
grO\\'
0 9).
from $270 million in 2013 to $490 million by 2018, at the rate of 12.40..{i
There is therefore strong interest in strategies that e<m enlumce the efficacy of
PGPR strains to improve agricultural productivity and reduce diseases due to bacteria, fungi,
nematodes and viruses.
[00185]
Our labs have conducted a comparative genom1cs study on our best-
performing POPR Bap strains, and
prest~nt
\Ve
were able to identify 73 genes that \Vere consistently
\Vithin all 28 genomes surveyed (including some strains published by other groups),
hut not present in other strains of R
activity (Fig. 2C} Importantly,
\Ve
amyloliquqf(~ciens
that \Vere known not to have PGPR
found that genes related to the uptake (exul) and
116 lization (uxuB) of pectin-derived sugars ( 18) \Vere alv./ays observed \Vithin these PGPR Bap
strains.
This led to our key hypothesis that pectin supplementation could increase root
colonization ability and metabolic activity of H amyloliquefaciens subsp. plantarum stn1ins,
thereby resulting in improved efficacy of these PGPR strains in terms of plant growth and
disease biocontrol.
[00186]
There
is
a significant knowledge base for pectin biochemistry and plant
hiosynthesis that cai1 benefit this project Henri Braconnot discovered pectin in 1825., and
pectin has been \vell charncterized as a major heteropo1ysaccharide of plant primary cell
\vaUs, in addition to cellulose and hernicellulose (20). For exmnple, the priTnmy cell wall of
56
WO 2016/054222
PCT/US2015/053239
Sycamore is composed of 34% pectin, 24% hemkel!u!ose, 23%1 cellulose, and l 9~o
hydroxyproline-rich glycoprotein (21 ). Pectin is at its highest levels \vi thin the fhJ1ts, leaves,
and roots of plants, so this is consistent \Vith the potential for pectin to provide a needed rootderived nutrient source for PGPR strains. Pectin is also found in the middle Jamel!a bet.,.veen
cells, \vhere it helps to bind cells together, and the availability and structure of pectin varies
arnong ph.mt species (22). Pectic materials are .frmnd in root hairs and have a thicker layer in
loamy soils than in sandy soils, and the duration of pectin in roots may depend on bacterial
enzymatic activity in the soil rhizosphere (23).
[00187]
Pectin degradation occurs through pectinolytk enzyrnes refen-ed to as pectin
lyases that are found in bacteria, fungi and higher plants (24). Bacteria are known to secrete
pectin lyases to degrade plant pectin. This patll\vay \Vas first reported in E. coli (25) and later
described in Bacillus subti!is (26), but besides our cmnparative genomics study (rnanuscript
in revie\v) and tw'o reports of Bap genome annotations (27, 28) there is no experimental
evidence for pectin utillzation by Bap strains or the ilnportance of pectin in their efficacy as
PGPR.
Pectinolytic
activity has
been
sh(nvn in the following bacterial genera:
Achromohacter, Atthrohacter, Agrobacterium, Bacillus, Clostridium, L'nvinia. l\·eudomonas.
and )(anthomonas (29, 30). Many of these bacteria are recogn1zed as plant pathogens, and the
degradation of pectin is a characteristic of soft rot disease a:s caused by Envinia spp.;
therefore, the competition for pectin as a nutrient source within plant rhizospheres could be
one of the mechanisms by \Vhich PGPR Hap strains antagonize plant pathogens \.vithout
themselves causing dmnage to plants. We have experhnental evidence that our sequenced Bap
strains encode and express a pectin lyase activity (Fig, 5), Additional experiments are needed
to detennine \vhether pectin lyase expression is a common trnh among all of our available
PGPR Bap strains (see Approaches).
[00188]
By producing and secreting pectin !yases, bacteria can degrade pectin and
uptake the pectin-derived sugars D-glucurnnate, D-gala,:turonate and D-mannose (31 ). The
sugars glucuronate and galacturonate can be taken up by bacteria via a hexuronate transporter
(end) system that encodes a hexuronate permease, From our comparative genomic study •.ve
have f()tmd that the exuT gene is conserved an10ng all sequenced PGPR Bap strains (n===28
57
WO 2016/054222
PCT/US2015/053239
with draft genome sequences). YVe subsequently designed a PCR pr1ll'.H,~r set targeting the exuT
gene in Bap strains and used this prin1er set to screen the 59 knmvn PGPR Bap strains in the
Auburn colkction (based on a phylogenetic analysis using both l 6S rRNA and ltrrB gene
sequences): of these 59 Bap strains,. 57 strains \Vere found to he PCR-positive for
~~xuT.
The
presence of this transporter among a large percentage of these strains supports the hypothesis
that pectin uptake is a cr1tical function for Bap strains. It is possible that the negative PCR
result for n:vo strains \:Vas a false negative (although this vvas repeated 3 times) or represents
exu T ge11es that are not recof,•11ized by the primer set. For th1s reason it is important to
evaluate actual utflization of pectin as a sole C source (see Objective 1 in Approach section).
[00189]
Besides pectin-derived sugar uptake, it is also critical for bacteria that utilize
pectin to have the degnH.lation pathway for using hexnronate sugars as a C and ener1:,1y source.
The uxuB Qene encodes a D-fructurnnaie oxidoreductase that is one of the enzvmes
~
~
responsible for degrading intracellular glucurnnate and galacturonate into 2-keto 3-
deoxygluconate (KDG) in E.coli and Fnvinia cfn:vsanthemi bacteria (32), and KDG is later
metabolized into pyruvate and 3-phosphogl:yceraldehyde. As \·Vith exuT, we identified uxuB as
a gene that is universally conserved among the 28 S(~qu~mced PGPR
recently screened the larger collection of 59 Auburn PGPR
Bap strains. Likewise,
\Ve
Bap strains for the presence of the
uxuB gene and found that 54 strains were found to be positive for uxuB. \\le do not know
i.vhether the 5 strains found to be PCR-negative for
uxuB lack the capacity for pectin
utilization as sole C source (see Objective 1 in Approach section). Identifying specific PGPR
strains that lack eh:her the capacity to uptake or use pectin as a sole C source will be helpful
because i.ve ca11 then use these strain(s) as a negative control that presumably would not
respond to the availability of additional exogenous pectin.
[00190]
Glucumnate, galactmonate and mmmose can he used as prinrnry carbon and
energy sources by bacteria such as Erwinia cluysanthemi (3L. 33). Since the acquisition of
carbon is essential for bi:K:terial energy generation and biosynthesis (34),
\'VC
hypothesize that
increased availability of pectin could promote the survival, persistence and n:ietaholic activity
of PGPR Bap strains \vithin plant rhizospheres, leading to improved plant growth and disease
control. Perhaps better PGPR efficacy is con-elated with the arnount of available pectin within
58
WO 2016/054222
PCT/US2015/053239
the roots or seeds of pardcnhff plant species? Iu this proposal \Ve
an~
focused on the 1ssne of
\.Vhether exogenously supplied pectin can induce better PGPR efficacy, and in subsequent
extramnral proposals we could explore the differences among plant species pectin content
(e,g., in Arabidopsis thaliana genetic mutants that lack pectin synthesis compared to \Vildtype plant pectin levels) and whether this correlates with PGPR efiicacy in promoting plant
gro\vth and disease control. There is only one report in the literature concerning the use of
exogenously supplied raw pectin to stimulate grovdh of plant-associated bacteria, in the case
of nitToge11-fixing
Azo.~piril/wn
isolates that had increased abundance in soils supplemented
\'-'ith pectin (35). However, in this research there was no attempt to evaluate any benefit to
plants in terms of growth promotion or disease control. Therefore, the amendrnent of pectin
along with a PGPR strain onto seeds or plant roots could be an innovative \vay to promote
plant gro\:i;.. . th and reduce disease, and reduce the variabillty inherem in field trfals of PGPR
strains.
[00191]
Rational and Siunificance
[00192]
There is a gn:.wving impetus to find sustainable means of irnproving animal and
plant health in agrk.ulture v{ithout resorting to the use of antibiotics. Recent federal initiatives
to reduce antibiotic usage and find alternative methods of improving animal and plant health
are highly synergistic ,.vith this research proposaL Prior to submission of federal funding
proposals, we need to test the hypotliesis tlmt pectin supplementation can increase the efficacy
of PG PR strains in inducing plant grmvth and disease controL This AAES proposal is
therefore essential in being_able to progress this research to a stage that it is competitive fix
extranmral support. There are nrnltjp!e ways that knmv!edge of the roles of pectin in bacterialplant interactions can benefit the practical application of PGPR strains:
1) The
supplementation of pectin onto seeds could e11hance PCIPR rhizosphere colonization which
could lead to rnore significant benefits in plant growth and disease reduction 2) The
persistence of PGPR strains within plant rhizospheres could be enhanced by periodic
rhizosphere drenches v,tith pectin, 3 .l Disease control mediated by PGPR strains occurs
through multiple mechanisms (direct antagonism, SAR and lSR), 'Nhich could be enhanced
59
WO 2016/054222
PCT/US2015/053239
through <.:alibrating PGPR and pectin inoculation timing to best reduce
dist~ase
pressure
fo.)111
plant pathogens.
[00193]
Approach
[00194]
Our experilnental approach will first focus on expanding ou.r kno\vledge of
pectin usage to all of the 59 PGPR Bap strains in the Auburn collection of Prof. Kloepper.
Our prelilninary PCR -based screen supports the hypothesis that pectin is
m1
irnportant source
of C and energy for the majority of PGPR Bap strains, and the experiments in O~jective l
will provide direct evidence f(ir pectin utillzation by these strains_ Importantly, Objective l is
also expected to indicate a fe\v Bap strains that do not have the capacity to utilize pectin. \Ve
predict that these pectin-incompetent strains wiU be at a competitive disadvantage for root
colonization,. and these strain(s)
\V.ill
then be important to include as negative controls in
Objective 3 for comparison vvith pectin-utilizing strains. To understand the genetic basis for
pectin-incompetence, in Objective 2 we will pick a representative Bap strain that is negatlve
in pectin lyase activity and/or pectin utilization as a sole C source (output of Objective I) for
genmne seque1icing and genomic comparisons \Vith other Bap strains. Thls will prov.ide
useful information on the specific genetic changes that have occurred in the small minority of
Rap strains that hKk the capacity to use pectin, aud indicate what other plant-derived
carbohydrates are being utilized in these strains. Most importantly, Objective 3 vviH test the
overarching hyJ)Othesis that supplementing pectin levels on seeds or in the plant rhizosphere
·will improve the efficacy of PGPR Bap strnins in stinrnlating plant grmvth and disease
control. V/e \:vil! use multiple Bap strains, including at I.east one strain that lacks the ability to
utilize pectin, and ·will apply a range of pectin concentrations as either seed amendn1ents or
rhizosphere drenches.
[00195]
Objective 1: Screen a large collection of Hap strains for pectin lyase activity
and pectin utilization as a sole C source.
[00196]
Hypothesis: The majority of Rap strains \Vil! express pectin lyase activity and
be able to use pectin as a sole carbon source.
60
WO 2016/054222
PCT/US2015/053239
[00197]
Methods:
[00198]
Objective t .1: Test Rap strains for pectin Ivase activity: Pt>t:tate-agm· media
'.viU be used to determine the pectin lyase activity of Bap strains. The bacteria \viii be grcn:vn
from cryostocks in the -80')C on Tryptic Soy Agar fTSA) at 28')C overnight, and an isolated
colony will be used to inoculate a Tryptic Soy Broth (TSB) 2 ml culture that will be incubated
at 28°C v..dth shaking at_250 rpm overnight A I ml aliquot of the overnight culture \.Vil! then
be p1petted hito a 1.5 ml microcentdfage tube and the bacterial cells \vi!1 he \Vashed three
ti1nes by subjecting the culture to centrifugation at 10,000 x g for 5 min and then re-
suspending the bacterial pellet in 1 x
phosphak~
buffered saline (PBS) and repeating the
process. To provide a uniform inoculum the bacterial re-suspension
l x PBS in a 2
1111
\Viii
be used to inoculate
culture tube and measun.' the turbidity of the bacteria! suspension '>'dth a
spectrophotometer until the Optical Density at 600 nm is approxiTnately 0..5. This
standardized bacterial suspension
\.Vill
be used to inoculate 20 microliters of suspension in
triplicate onto a pectate-agar 1nedimn (36) used for detennination of pectin lyase activity. The
pectate-agar medium plates \\dll be incubated at 28°C overnight and then a 1~(i cetyltrimethyl
ammonium bromide (CTAB)
\),-'ill
be poured over the surface of the plate, with a ckar zone
present around the bacteria! colony (Vig. 5) forming after 30 min indicating pectln !yase
activity. The magnitude of the zone of clearing will be measured in n1m and recorded in an
Excel spreadsheet, \.Vith average zones of clearing being determined for each of the Bap
cultures.
[00199]
Objective 1.2: Test Bap strains for the use of pectin as a sole C source: The
capacity of each of the 59 Bap strains to utilize pectin as a sole C source will be assessed
using a minimal Tris-Spizizen salts (TSS) medium (37) as the base medium supplemented
with 1% pl.ant pectin (citrns source). Each of the bm.:terial cultures \.v:il! be prepared as for tlw
pectin !yase assays 'with bacterial suspensions \.Vashed in I x PBS, normalized to an ODoxi '''
0.5, and then 100 microliters of a L too dilution \\'i!I be used to inoculate 1.9 ml TSS +
1°-'~
pectin cultures, in triplicate. Broth cultures \vill be incubated at 28('C with shaking and OD61i>
readings
\Vil!
be taken over a 36 hour time period, In a preliminary experiment, \Ve have
61
WO 2016/054222
PCT/US2015/053239
observed that two of uur previously
sequenc{~d
PGPR Bap strains, AP 143 and AP 193, could
utilize pectin as a C source for grrnvth (Fig. 6), As a negative control, a non-PGPR Bacillus
thuringiensis sufop. kurstaki strain HD73 \Vas obtained frorn the USDA ARS culture
collection (Ames, Imva) that \Vas identified as a non-pectin utilizing strain based on its
genome sequence. This strain \Vas not observed to grmv using pectin as a sole C source (Fig,
6).
[00200]
Potential Outcomes: Bap strains that have been confirmed to contain exu T and
uxuB genes vd!l express pectin lyase activity and grmv using pectin as sole C source. Some
strains v.lill show a larger zone of clearing in the pectin lyase experin1ents, and any highexpressing strain(s) \Vil! be used in subsequent experiments.
[00201]
There may he strains that do not grmv using pectjn as a sole C source, and one
of these strains can be used in Objective 2 for comparative genomics and in Objective 3 as a
nel!ative
control.
....
[00202]
Potential Problems: It is possible that despite hav.ing the necessary genes a
strain may not phenotypica!!y express pectin lyase activity or pectin utilization under in vitro
conditions. Based on preliminary results \\>e do not expect this to be a problem, and knO\·V that
,.ve have a !east hvo strains, AP143 and AP19J, that were previously identified as top-
performing PGPR strains, \Vere subjected to genome sequeiKing and have been shtW.ill lO
degrade and use pectin as a C source.
[00203]
Ohjecth•e 2: Conduct a comparative genonuc analysis that includes Bap
strnin(s) that lack the capacity for pectin utilization
[00204]
Hypothesis: A Rap strnin(s) unable to use pectin \.vill have geno1rnc
reammgemt~nts
[00205]
in
tht~
pathways responsible for pectiu degradatiou and./or utilization.
hfothods:
62
WO 2016/054222
[00206]
PCT/US2015/053239
Obit!Ctive 2, 1: Determine genome seguem.:e for strain(s) lacking_ 12ectiq
utilization: Based on the results from Objective 1, \Ve \Vil1 select Bap strain(s) for genome
sequencing that do not have any evidence for pectin de.gradation andior pectin utilization.
Based on the results of PCR-based screening of e),·uT and uxuB genes (see above), we do
expect a fo\V strains to be negative for pectin utilization as a sole C source. Each strain wiH be
grown in medium-scale (-· 100 inl TSB) at 28"C until the culture reaches an OD600 of 0. 5-0.8.
Genomic DNA isolation
\Vil!
be conducted using a harsh bead-beating method (l\foBio,
Cmlsbad, California) in order to achieve sufficient yield of gDNA from gram-positive
bacteria. The gDNA ,,viii he quantified using a Qubit t1uorometer. If \Ve achieve > 50 ng of
high-quality gDNA for each strn:in, then
\VC \Vilt
use a Nextera tagmentation kit (Il!umina, San
Diego, CA) for preparation of bar-coded sub-libraries (otherwise, the Nex tera XT kit
\Vill
be
adopted v./hich can use as little as l ng gDNA). We \.vill include no more than 5 strains per
lllurnina MiSeq sequencing run (available in the Rouse Life Sciences building), and use 12
piVI final concentration of the pooled bar-coded libraries along ,.vi th l ~--0 spike-in of the PhiX
internal control. The sequence daia
\Viii
be exported in .fastq fbrrnat frmn the MiSeq and
imported into the CLC Genomics Workbench (CLC hio, Cambridge, ivlA). Vv'e
,,,.m
trim
sequences for quality (trim sett:ing 0.01), followed by de novo assernbly. If large contigs do
not result from the assembly, we will then evaluate different assernh!y criteria using CLC
Genomics, Ray, and VelvetOptimiser assemblers., in order to identity the largt.$t contigs
possible for each strain. Based on our prevfrms experience with next-gen sequencing and
assembly of bacterial genomes, \Ve expect excellent results \Vhen >80x coverage is attained.
[00207]
Objective 2.2: Analvze genomes for pectin- and carbohvdrnte utilization
patlnvavs: The bacteria! genome sequences wiH be annotaied by exporting the entire set of
contigs for each strain into the RAST server (http://rast.nmpdLt)rg/).
RAST output 'Nill
indicate the presence of pred:icted carbohydrate utilization patlnvays. In addition, \Ve
\Vill
manually compare the cornplete set of genome contigs for each strain \Yith the genes required
for pectin lyase (pe!B), hexuronate transport (exu1) and hexuronate utilization (uxuB)
obtained from Bap strain AP193 using pairwise tBLASTx comparisons al NCBI. These
analyses will indicate the presence of specific genes if they are present in a genome even if
63
WO 2016/054222
PCT/US2015/053239
the previous PCR-based screening \\'as ineffec.:tive because of gene
sequ~mce
differences at
the primer annealing site, Fmthennore, \Ve \viU compare the genomic archltecture of each
non-pecdn ntihzing strain \Vith that of AP 193 and other sequenced Bap genomes to query the
local region surrounding each pectin uti !ization pathway. This could indicate \Vhether the
nature of the defect in pectin utilization is related to transcriptional regulation (change in
promoter region}, point mutation(s) in specific genetic loci, or cornplete genetic deletion or
rearrangement of the pectin utilization pathwa::,ts.
[00208]
Potential Outcomes: A draft genome for Bap strain(s) that lack the capacity to
degrade and/or utilize pectin, and annotation of the genes and genotnic regions involved in
each patlnvay.
[00209]
Potential Problems: \li/e have considerable experience conducting !vbSeq
sequencing nms (>20 at Auburn), but there can be variability concerning the sequencing yield
obtained per straiJL To 1nitigate that we \'•iill aim to achieve at least lOOx genome coverage on
the strain, and also budget for nvo sequencing nms in case there is any problem \Vith the first
sequencmg nm.
[00210]
Objective 3: Evaluate pectin supplermmtation along v.,rith Bap strains lbr
soybean growtl1 prornotion and disease control
[00211]
Hypotl1esis: Supplementing pectin levels on soybean seedlings will improve
the efficacy of PGPR Bap strnins in stimulating pl.ant blTOwth and disease control.
[00212]
fvkthods:
[00213]
Objective 3J: Determine pectin dose-dependent enhancement of PGPR strain
growth:
[00214]
It is important to first det.ennine the concentration of pectin to use in
subsequent plant grm:\irth experiments. In order to track strain colonization, \Ve V>..-ilJ select for
rifampicin-resistant mutants (50 p.g/ml Rit) on TSA and select three independent RifR mutants
64
WO 2016/054222
PCT/US2015/053239
per strain to use as replicates. Soybean seeds (Park Seed, Hodges, SC)
\Vil!
be so\vn in
Styrofoam trays and three \Veeks after planting seedlings \:vill be transplanted into a 4.5 inch
squme pot \Vith a i;.:ommercial potting substrate (Sunshine rnix, Sun Gm Horticulture,
Aga\vam, \.faine), Three days after transplanting, seedlings \Vill he drenched with lO ml of
6
either AP193 or HD73 spores (10 CFU.iseedling) applied in 1) sterile \Vatec. 2) 0.01% {\v/v)
pectin, 3) 0 . 1i~.··(i pectin, or 4) .1 ih1 pectin, \·Vith each of the 8 treatment groups (2 strains x 4
pectin amounts) in quadruplicate for 32 total pots. Plants \Vil! be transforred to the greenhouse
and watered daily for 21
da).tS.
Pectin supplementation at the indicated concentrations will be
conducted at ,.veek!y intervals . J\t 2 l days post-inoculation, 10 g of rhizosphere soil will be
sampled from each of the pots, and the dry shoot and root weights 'viH be detennined. The
1
soil 'vill be homogenized in 90 inl of sterile \vater (HY dllution) and then serially diluted to
6
10-6 dilution and each of the dilutions from 1ff1 to 10- v.dll be plated_onto TSA plates In order
to detennine the mnnber of RifR colony fanning units (CFU)/g of soil. The results of this
experiment are expected to shm.v a significant pectin dose-dependent increase in strain AP 193
CFU/g of soiL, ·with no significant increase observed fi)r pectin-incompetent strain HD73. The
!O\vest dose of pee.tin showing the strongest induction \Vil1 be used in Obj. 3 2.
[00215]
Potential Outcomes: Determination of tbe pectin concentration(s) that result in
increased grov;..-th of the PGPR Bap strain \\.>hen amended into a potting substrate.
[00216]
Potential Problems:
ldt~aHy
this experiment would be conducted usmg an
agricultural soil; ho\vever, it is exceedingly diJTicult to discriminate het\veen the indigenous
rhizosphere microbiota and the inoculated PGPR strain in soils, We selected a potting mix
because this is vd1at is conunonly used for greenhouse grown crops,. so it is relevant to
conditions used by commercial grm.vers, \vhile also allm.ving the more facile recognition of
the PGPR strain \vhich has a unique colony morphology. To confin:n that the CFC counts are
due to the inoculated PGPR Bap strain AP 193, representative colonies \viii be selected to
conduct PCR using an AP l 93 stra:in-specific PCR primer set that has been developed in the
Lites lab (data not shO\'lin).
65
WO 2016/054222
[00217]
PCT/US2015/053239
Obit!ctive 3.2: Evaluate i:;iectin-iuduction of PGPR-mediated ulant gro\\·th
promotion:
[00218]
Soybean seedlings viiiH be will be smvn into larger 8 inch pots that contain a
sandy loam soil potting soil to \.Vhich 497 mg phytate is added per kg, It has previously been
shO\vn that Bap strnin FZB42 can mediates its plani gnJ"\.vth-promoting effects in part via
phytase activity, and that the addition of phytate to soils can help to observe this PGPR-
mediated plant grtwvth-pronwting effect (38). The treatment groups ,,,,,,.ill. include l) no spore
treatment, 2) !06 CFU/seedling PGPR Bap strain APl93 and 3) 106 CFU/seedling of one
other PGPR Hap strnin that shows strong pectin lyase activity (from
seedlings in each treatment group
Ol~jective
l). The
be drenched two days atl.:er transfer to the greenhouse
\Viii
with either sterile \\'ater or pectin (at a 0.. .;) to be determined above), using ten replicates \.vith a
completely randmnized design There \\. ill therefore he 6 treat1nent gronps x lO replicates==== 60
1
total pots. Pectin addition \Vill occur at I \veek intervals and plants \\"ill be grown for 4 \veeks.
Fresh and dry weight of root and shoot will be rneasmed at the cmnpletion of the greenhouse
experiment Root morphology vvill be analyzed by root scanner. The fresh and dry \Veight of
shoot and root variation of each treatment \.viH be con1pared and anal'yzed using ANO'v'A at
5~"ii
level of significance (SAS 9.1 sofi.ware).
[00219]
Potential Outcomes: The addition of pectin to Hap-treated soybean seeds \v111
result in an increase in soybean root and shoot grmvth relative to non-pectin-treated plants.
No pectin-mediated increase will be observed in the plants \Vithout PGPR strain inocu!um.
[00220]
Potential Proh!erns: There can he m1fbreseen issues, such as a disease
outbreak, in experimental plants. V./e \vill take ample precautions to 1imit disease incidence,
and plan to repeat this experiment at least
one'-~
and more if necessary. The t.irning of the
experiment may need to be extended to achie..,;1e significant differences among treatment
groups. It might be useful to include other crop species (in extramural proposals), since pectin
content cai1 vary il1 different plant species and pectin supplementation might be more useful
in plants limited in pectin content
66
WO 2016/054222
[00221]
PCT/US2015/053239
Obit!ctive 3.3: Evaluate Qectin-induction of PGPR-mediated giant disease
control:
[00222]
We will evaluate pectin-induced plant disease control us1ng
pathogens:
1-~rthium
t\.'<iO
difterent
u!timurn that causes pre- and post-emergent seedling damping-off, and
Rhizoctonia solani that causes root rot and hypocotyl !.esions on cucurnber and soybean,
respectively. Hence, t\vo separate hrroups of experiments
\Vill
be completed, one with each
6
pathogen. The PGPR Bap strains '""ill be applied as l 0 CFU spores, a comrnon rate used i,vith
PGPR as commercial seed treatments, to each cucumber or soybean seed in a Styrofoam tray,
and a drench of 10 ml ofpet:tin at the dose used in Experiment #2 \vill be applied at time zero.
\Ve wit! include at least t\VO PGPR Bap strains that are know11, from previous \vork in
Klot~ppt~r's
lab, to inhibit Pythium damping-off aud Rhi::octonia root rot and v..tere positive for
pectin utilization in Objective
#C along '>'Vith tw·o negative contxols: a non-PGPR strain HD73
and a pectin-incompetent PGPR strain identified frorn Objective #1 and sequenced in
Objective #2. Therefore, each experirnent will contain 8 treatinents: 2 PGPR strains
previously shmvn to control the respective pathogen and t\vo negative control strains, with
and 'Without pectin supplementation. We vvill use ten replicates and the
t~xperiments \Vill
be
conducted in a completely rn11domized design. After 3 days and prior to plant gem1ination,, a
l~vthium
u/timwn or Rhizoctonio sokmi culture, respectively, gro\vn on potato-dextrose agar
\'-'ill he homogenized ,,vithin
the agar, diluted \v.ith sterile \Vater and homogenized further, and
6
then used to inoculate seedlings \vith approx. l 0 cells/ ml in 10 ml. Inoculated plants ·will be
placed into a dew chamber at 10oih) hurnid.ity in the dark fbr 2 days at
24'~C
and then
tra11splanted into a pot aud transforred to the greenhouse, Plants '>Vill be ·watered daily and
mcmitored for damping off At 7 and 14 days post-pathogen inoculation the plant mortality
count \Vill be determined for each treahnent group, and these data
\Viii
be compared and
anal'yz.ed using ANO VA at 5ci/o significance kvel (SAS 9.1 ).
[00223]
soybean seeds
Potential Outcomes: The addition of pectin to Rap-treated cucumber and
\Vil!
result in enhanced protection of PGPR strains against l)'thium ultimum
and Rhlzoctonia so!ani-mediated damping off, respectively.
67
WO 2016/054222
[00224]
PCT/US2015/053239
Pott~ntfal
Problems: There ai-e several variables that could impact this
experiment, in particular the timing of the inoculation of PGPR, pectin, and pathogen on the
cucumber and scrybean seed.lings. Depending on the results of the experiment, we may need to
change tl1e timing and/or concentrntjon of these factors in repeating this experiment. A !so,
because these pathogens induce damping off at an early point in the plant lifo cycle, there may
be snfiicient numbers of metabolicaUy active Hap cells present at early time points such that
pectin amendments are less necessary. If this is observed then we may consider the use of an
alternative pathogen i;.:hallenge
[00225]
References
[00226]
L
\\'itl1
more mature cucumber or soybean plants.
Kuehn Bl'vL 2012. FDA aims to curb fam1 use of antibiotks. JAJvV\
the journal of the American :tvledical Associatkm 307:2244-2245.
[00227]
2.
Raupach GS, Kloepper JW. 1998. !vhxtures of pl.ant grovvth-promoting
rhizobacteria enhance biological control of multiple cucumber pathogens, Phytopathology
88: l.158- 1164.
[00228]
Adcsemoye AO, Torbert HA, Kloepper J\V. 2009. Plant :!$'Towth-
promoting rhizobacteria allow reduced application rates of che1nical fortilizers. rvlicrohial
ecology 58:921-929.
[00229]
4.
Yan Z, Reddy rvlS, Ryu C:rvL lVIcinroy JA, \Vilson r-vl, Kloepper JW.
2002. foduced systemic protection against tornato late blight elicited by plant grmvthpromoting rhizobacteria. Phytopathology 92: 1329- l 333.
[00230]
5.
Ryu Cf\·1, i'vlurphy JF,. Ivlysore KS, Kloepper JW. 2004. Plant growth-
promoting rhizobai;.:teria systemically protect Arnbidopsis tha!iana against Cucumber mosaic
virus by a salicylic acid and NPR 1-independent and jasmonic acid-dependent signaling
path\vay. The Plant journal : f(_)r t:cll and molecular biology 39:3 81-392.
68
WO 2016/054222
[00231]
6.
PCT/US2015/053239
Compaore CS, Nielsen DS, Smvadogo-Lingani H, Berner TS,
Nielst~n
KF, Adimpong DB, Diawara B, Ouedraogo GA, Jakobsen 1v1, Thorsen L 2013. Bacillus
amyloliquefacit~ns
ssp. plamarum strains as potential protective starter cultures for the
production of Bikalga, an alkaline fonnented lhod. J Appl !V:licrobiol 115: 133-146.
[00232]
7.
Avdeeva LV, Dragovoz IV, Korzh Iu V, Leonova NO, lutinskaia GA,
Berezhnaia AV, Kuptsov VN, i'vfondrik
~-JN,
Kolomiets EL 2014, [Antagonistic activity of
Bacillus amy!o!iquefaciens subsp. phmtarum Il'v1V B-7404 and Bl.l\.'1 B-439D strains to\vards
pathogenic bacteria and micrnrnycetesj. \.likrohiol Z 76.:27-33.
[00233]
Klot~ppt~r
8.
J\V,
Ran C, Carrias A, vVi!liams l\tA, Capps N, Dan BC, Ne\:vton JC,
Ooi EL, Bro\vdy CL, Terhune JS, Ules tv1K 2012. Identification of Bacillus
strains for biological control of catfish pathogens. PloS one 7:e45793.
[00234]
9.
Calvo P, Nelson, L & Kloepper, J.W. 2014. Agricultural uses of plant
biostimulants. Plant Soil.
[00235]
10.
Basha11 Y, de-Bashan LE, Prnbhu SR, Hernandez JP. 2014, 1\dvances
in plant gro\vth-promoting bacterial inoculant tedmology: fi.mnulations and practical
perspectives {1998-2013 ). Plant Soil 378: 1-33.
[00236]
11.
Z.,a1.d.i ,S,. F«
., ,,mg
S. )!1 BR
I '"::J
"' )06.
- . S.
_._.mam. S,
. , uMusarrat ....
, 1gm"ficance of
Bacillus subtilis strain
SJ~ tol
as a bioinoculant for concurrent plant grmvth promotion and
nickel accunrnlat.ion in Brassica juncea. Chemosphere 64:991-997.
[00237]
12.
He PF, Hao K, B1on1 J, Rueckert C, Vater J, \fao ZC, Wu YX, Hou
!\.-:IS, He PB, He YQ, Borriss R. 2012. Genome sequence of the plant grm.vth promoting strain
Bacillus amyloliquefadens snbsp plantarum 89601-'{2 and expression of rnersacidin and
other secondary metabolites, Journal of bioteclmo!ogy 164:281-291.
[00238]
13.
\.fariappan A, t\..fakare\vicz 0, Chen XH, Borriss R. 2012. Tvlo-
Component Response Regulator DegU Controls the Expression of Bacilysin in Plant-Gro\vth-
69
WO 2016/054222
PCT/US2015/053239
Promoting Bacterium Bacillus amylohquefaciem.; FZB42. Journal of molecular microbiology
and biotechnology 22: 114-125.
[00239]
14.
Blom J, Rueckert C, Niu B, Wang Q, Burriss R. 2012- The Complete
Genome of Bacillus amyloliquefaciens subsp plantarum CAU B946 Contains a Gene Cluster
forNcmribosomal Synthesis ofHrnin A. Journal ofbacterio!.ogy 194:1845-1846.
[00240]
15.
Raupach GS, Kloepper J\V. 1998. 1Vhxtnres of plant grm;s, t.h-pron10Hng
1
rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology
88: 1158-1164.
[00241]
16.
A!tinok HH, Dikilitas !VC Yildiz HN. 20 ! 3. Potential of Pseudomonas
and Bacillus Isolates as Biocontrol Agents against Fusarium \:Vilt of Eggplant Biotechnol
Biotec Eq 27:3952-3958.
[00242]
17.
Kloepper J\V, Ryu Cl'vf, Zhang SA. 2004, Induced systemic resistance
and prornotion of plant gnYwth by Bacillus spp. Phytopathology 94: 1259-1266.
[00243]
18.
Kloepper JW_
1994. P!ant-gro,vth-prnrnoting rhizobacteria (other
systems), p. 139 - 154. In Okon ''{ (ed.), Azospirillu.m.iplant associations. CRC Press, Boca
Raton.
[00244]
19 .
Anonymous 2013, posting daw. Biostinrnlants market - By Active
Ingredients, Applkafams, Crop Types & Geograplxy ······· Global Trends & Forecasts to 2018.
[Online.]
[00245]
20.
Anonyrn.ous, posting date. Plant Cell walls. University of Georgia,
21.
Darvlll A, tvkNeil J\1, Albershefrn P, Dehner D. 1980. The pritnary cell
USA [Online.]
[00246]
\'-'alls offlmvering plants. The biochemistry of plants L91-162.
70
WO 2016/054222
[00247]
22.
PCT/US2015/053239
Srivasta'va P, Malviya R. 201 I. Smm.:es of pectin, extraction and its
applications il1 pharmaceutical industry-An overview. Indian journal of natural products and
resourct$ 2: l 0- 18.
[00248]
Hm:ve CG. 1921. Pectic material in root hairs. Botanical gazette:3 l3-
320.
[00249]
Nmnasivayarn E, Ravindar J, !vlariappan K, Jiji A. Kurnar ?vL. Jayaraj
RL. 2011. Production of extracellular pectinase by Bacillus cereus isolated from inarket solid
'\vaste. J Bio.anal Biomed 3:070-075.
[00250]
Ash\vell G, \:Vahba AJ, Hkkman J. 1960. Uronic add nwtabolism in
bacteria L purification and properties of uronic acid isomerase in escherichia coli.. Journal of
Biological Chemistry 235: 1559-1565.
[00251]
26.
Kloepper J, :tvktting Jr F. 1992. Plant growth-promoting rhizobacteria
as biological control agents. So.il rnicrohial ecology: applications in agricultu.ra1 and
envirnmnental management:255-274.
[00252]
,.
')'1
'-
Niazi A, Manzoor S, .Asari S, Bejai S, Meijer l, Bongcam-Rudloff E.
2014. Genome analysis of Bacillus amyloliqudli.ciens Subsp. plantarurn UC\1B5ll3: a
rhizobacterium that improves plant gro\vth and stress management. PloS one 9:el 04651.
[00253]
28.
Chen XH, Koumoutsi A, Scholz FL. Eisenreich A, Schneider K,
Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva 0, Junge H, Voigt B, Jungblut PR,
Vater J, Sussnrnth R, Liesegang H, StrjHmatter A, Gottschalk G, Borriss R. 2007.
Comparative analysis of the complete genome sequence of the plant grO\.vth-promoting
bacterium Bacillus amyloliquefac.iens FZB42. Nature biotechnology 25: 1007-l 014.
[00254]
29.
Rombouts FM. 1972. Ckcurrence and properties of bacterial pectate
!vases. Landbouwhogeschoo!
te
.......
.,/
\~lageni1rnen
.
......
.
.......
71
WO 2016/054222
[00255]
30.
PCT/US2015/053239
Voragen AGJ. 1972. Characterization of pt~ctin lyases on pectins and
methyl oligogalacturonates.
[00256]
31.
l\.'1ekjian KR, Bryan
E~.f,
Beall B\V, Moran CP. 1999. Regulation of
Hexu.ronate Utilization inBaciBus subtilis. Jouma1 of bacteriology l 81 :426-433.
[00257]
32.
Hugouvieux-Cotte-Pattat N, Nasser W, Roben-Baudouy J.
1994.
Molecular charactedzation of the En:vinia chrysanthemi kdgK gene involved in pectin
degradation. Journal of bacteriology 176:2386-2392.
[00258]
33.
Hugouvieux-Cotte-Pattat N, Robert-Baudouy J.
l 987. Hexuronate
catabolism in Erwinia d1fysanthemi. Journal of bacteriology ! 69: 1223-123 l.
[00259]
34.
Kenneth T Nutrition and Grmvth of Bacteria, p. Ol-06, online textbook
of bacteriology Online textbo<Jk.
[00260]
35_
Mehanni l\Hvfal\.-t, JL\_ 2012, p 99 - 11 l. Egypt Journal of Botany 2nd
International Conference, rvlinia. University.
[00261]
36.
Kobayashi T, Koike K, Yoshimatsu T,. Higaki N, Suzumatsu A, Oza,va
T, Hatada 'f, Ito S. 1999. Purification and propenit~s of a 1ow-molecu1ar-\ve1ght, high-alkaline
pectate !yase from an alkaliphilic strain of Bacillus. Bioscience, biotechnology, and
biochemistry 63:65-72.
[00262]
""7
.::>1
Shingaki R, Kasahara Y, lwano 1\.·1, Kuwano M, Takatsuka T, lmme T,
Kokeguchi S, Fukui K 2003. Induction of L-form-like cell shape change of Bacillus subtllis
under microculture conditions. rvncrobiology 149:2501-2511.
[00263]
38.
Ramirez CA, Kloepper JW. 2010. Plant grO\vth pronmtion by Bacillus
amyloliquefaciens FZB45 depends on inoculmn rate and P-rdated soil properties. Biol
Soils 46:835-844.
72
Ft.~rt
WO 2016/054222
[00265]
It
\Vill
PCT/US2015/053239
be readily apparent to one skilled in the art that varying substitutions
and modifications may be made to the invention disclosed herein \Vithout departing from the
scope and spirit of the invention. The invention 11lustratively described herein suitably may be
practiced in the absence of any element or elements, lilnitation or limitations \vhich is not
specifically disclosed herein. The tem1s and expressions ·which have been employed are used
as terms of description and not of limitation, and there is no intention in the use of such terms
and expressions of excluding any equivalents of the features shown and described or portions
thereof: but it is recognized that various modifications are possible Vidthin the scope of the
invention. Thus, it should be understood that although the present invention has been
illustrated by specific embodiments and optional foatures, modification and/or variation of the
concepts herein disclosed may be resorted to by those skilled in the art, and ihat such
modifications and variations are considered to be \Vithin the scope of this invention.
[00266]
Citations to a number of patent and non-patent references are made herein.
The cited reforences are incorporated by reference herein in their entireties. In the event that
there is an inconsistency bet\veen a definition of a term in the specification as compared to a
definition of the tern1 in a cited reference,
th"~
definit1on in the specification.
73
tenn should be interpreted based on the
WO 2016/054222
PCT/US2015/053239
CLAIMS
\'Ve claim:
1.
An inoculant comprising: (a) a plant e,Yfmvth promoting rhizobacteria
{PGPR) that expresses a protein associated \vith pectin metabolism and (b) a saccharide
comprising pectin or a pectin-related saccharide.
2.
The inoculant of claim l, \vherein the PGPR is a Bacillus species.
3.
The inoculant of claim 2, \.Vherein the Bacillus species is Bacillus
amyloliquefaciens subspecies plantarurn.
4.
The inoculant of clain1 I, \vherein the pectin-related saccharide
comprises hydrolyzed pectin,_ D-galacturonate, D-glucuronate, or mixtures thereof.
5.
The inoculant of claim l, wherein the saccharide is a polysaccharide
comprising D-galact.uronat.e monomers.
6.
The inoculant of claim I, \Vherein saccharide is a heteropolysaccharide
comprising D-galacturonate monomers \.vhich represents at least
sm·o of all monomers of the
heteropolysaccharide.
7
The inoculant of claim 6, wherein the heteropolysaccharide further
comprises one or more monomers selected from D-xylose, D-apiose, and L-rhamnose.
8.
The inocu1ant of daim l , •vherein the saccharide is a heterogeneous
mixture of polysaccharides or monosaccharides cornprising D-galacturonate monomers or Dglucuronate monomers, and the sum of D-galacturonatc rnonomers and D-gfocuronate
monomers in the mixture represent at least soih1 of the total monomers of the rnixture.
9
Plant seeds coated with the inocu!ant of claim 1.
10.
Animal teed cmnprising the inoculant of c !aim l .
74
WO 2016/054222
lL
PCT/US2015/053239
.A
1rn.:~thod
for improving plant gn)\·vth or phml health, the method
comprising: (a) treating plants, seeds,. or soil \Vith a plant grmvth promoting rbizobacteria
(PGPR) that expresses a protein associated \.vith pectin metabolisrn and (b) treating plants,
seeds, or soi! \.vi th a sacdmride comprising pectin or a peciin-related saccharide.
12.
The method of claim 11, \Vherein the plants, seeds,. or soil are treated
concurrently 11vith the PCiPR and the saccharide.
13.
The method of claim l ! , wherein the plants, seeds, or soil are treated
first \.vith the PGPR and subsequently the plants, seeds, or soil are treated with the saccharide,
14.
The method of da:im "I l, \.vherein the plants, seeds, or soil are treated
first \vitl1 the saccharide and subsequently the plants, seeds, or soil are treated \vi th the PGPJC
15.
The method of claim l l, \vherein the method improves plant grmvth or
plant health by controlling soil-borne pests selected from nematodes and insects.
16.
The method of claim l l, wherein the method improves plant grmvth or
plant health by controlling a disease selected from a bacterial disease, a fungal disease, and a
viral disease.
17.
The method of claim l l, \.vherein the PGPR is a Bacillus species.
18.
The method of claim l 7, wherein the Bacillus species is Bacillus
a.myloliqlH;'.fhciens subspecies p!antarum.
19.
The method of claim l L, \Vherein the pectin-related saccharide
comprises hydrolyzed pectin, D-galacturonate, D-g!ucuronate, or mixtu.res thereof.
20.
The method of dairn 11, \vherein the saccharide is a polysaccharide
comprising D-galacturonate monom.ers.
75
WO 2016/054222
2L
PCT/US2015/053239
The method of clalir1 I. l, \Vherein saccharide is a lwteropolysaccharide
comprising D-galacturonate monomers \\'hi ch represents at least 500.-·o of all monomers of the
heteropolysa<.:charide.
22.
The method of da:im 21, wherein the heteropo!ysaccharide further
comprises 011e or more n10nomers selected 11:om D-xylose, D-apiose, and L-rhamnose.
The method of claim 11, \vherein the sacdrnride is a heterogeneous
mixture of polysaccharides or monosaccharides c01nprising D-galacturonate monomers or D~
-
glucuronate monon1ers, and the sum of D-galacturnnate monomers and D-glucuronate
~
~
monomers in the mixture represent at least 50°,{1 of the total monomers of the mixture.
24.
!\ method for irnproving anin1al growth or aniinal health,. the method
comprising: (a) administering to animals a plant gn_)\Vth promoting rhizobacteria (PGPR) that
expresses a prntein associated vdth pectin metabolisn1 and (b) administering to animals a
saccharide comprising pectin or a pectin-related sact:haride,
25.
The method of claim 24, wherein the animal are administered the
PGPR and the saccharide concurrently.
26.
The inethod of claim 24, wherein the animals are administered first the
PGPR and subsequently the animals are administered the saccharide.
27.
The method of claim 24, \:1.rherein the animals are administered first the
saccharide and subsequently the animals are administered the PGPR.
28.
The meth1)d of claim 24, whereiu the PGPR is a Bacillus species.
29.
The method of claim 28, \Vherein the Bacillus species 1s Bacillus
amyloliqut.:f<.:u.:iens subspecies plantarum.
30.
The method of claim 24,. \vherein the pectin-related saccharide
comprises hydro!:yzed pectin, D-galacturnnate, D-glncmona.te, or mixtnres thereof.
76
WO 2016/054222
3L
PCT/US2015/053239
The nwthod of claim 24, \vherein
tht~ saccharidt~
is a polysaccharide
comprising D-ga!acturonate monomers.
32.
The method of claim 24, wherein saccharide is a heteropolysaccbaride
comprising D-galacturonate monomers vvhich represents at least
500.··~
of all rnonomers of the
heteropol.ysaccharide.
33.
The method of cla.irn 32, \vherein the heteropolysaccharide further
comprises one or more monomers selected from D-xylose, D-apiose, and L-rhamnose.
34.
The method of claim 24, vvherein the saccharide is a heterogeneous
mixture of polysacdmrides or monosaccharides cornprising D-galacturonate monomers or D-
glucuronate monorners, and the sum of D-galacturonate monomers and D-g!ucuronate
monomers in the mixture represent at least 50~>o of the total monomers of the mixture.
77
:.;;
0
N
0
r-
100
100
100
I
100
"'c
"'=i
OJ
81
-I
c
-I
100
m
"'mm:::c
-"'
-I
;;c
c
0.1
r
m
a>
FIG. 1A
CEREUS ATCC 14579
PUMILUS INR?
PUMILUS SFR032
LICHENIFORMIS DSM13
SONORESIS KCTC 13918
ATROPHAEUS 1942
MAJAVENSIS KCTC 3706
I
L.1QQ....
TEQUILENSIS KCTC 13622
B. SUBTILIS SUBSP. SUBTILIS STR. 168
AP254
B. SIAMENSIS KCTC 13613
B. AMYL. LL3
100
} B. AMYL. SUBSP.
B. AMYL. DSM?
AMYLOLIQUEFACIENS
B. AMYL. TA208
96
98 AB01
B. SUBTILIS GB03
100
BACILLUS SP. 586
97
B. AMYL. FZB42
B. AMYL. CAU 8946
,-B. AMYL. YAU B9601-Y2 ~ B. AMYL. SUBSP.
AP 193
PLANTARUM
AP143
I__ B. AMYL. AS433
AP71
AP79
a-,
"""
0Ut
B.
B.
B.
B.
B.
B.
B.
""'
N
N
N
liQQt
--""""""
0
""d
(""l
g
00
N
0
"""
Ut
0Ut
~
N
~
l.O
:.;;
0
N
0
a-,
"""
I
r------1.:....:.:0~0
{
100
I
100
-......:....::~
1001
100
I
100
I
"'c
"'=i
OJ
100
-I
c
-I
m
"'mm:::c
-"'
-I
;;c
c
-------0.1
r
m
a>
FIG. 1B
1001
0Ut
B. CEREUS ATCC 14579
B. PUMILUS INR?
B. PUMILUS SFR032
B. LICHENIFORMIS DSM13
B. SONORESIS KCTC 13918
B. ATROPHAEUS 1942
- B . MAJAVENSIS KCTC 3706
B. TEQUILENSIS KCTC 13622
AP254
B. SUBTILIS SUBSP. SUBTILIS STR. 168
B. AMYL. DSM?
} B AMYL SUBSP
I 100
B. AMYL. TA20S
AMYLOLIQUEFACIENS
100
---=-=--=---- B. AMYL. LL3
100 - B. SIAMENSIS KCTC 13613
- B. AMYL. CAU 8946
-B. AMYL. YAU B9601-Y2
100
AB01
100
BACILLUS SP. 586
100
B. SUBTILIS GB03
B. AMYL. SUBSP.
-AP143
PLANTARUM
AP193
100
B. AMYL. AS433
B. AMYL. FZB42
45
100 ~AP79
AP71
""'
N
N
N
--"""
N
0
""d
(""l
g
00
N
0
"""
Ut
0Ut
~
N
~
l.O
WO 2016/054222
PCT/US2015/053239
3/10
S3cmrv3.:I ~31SA.ssns .:10 CJ3s~nN
SUBSTITUTE SHEET (RULE 26)
:.;;
0
~BACILLUS
en
100%
w
~
(B)
:::::>
~
00%
u..
00%
w
~
w
en
>en
:::::>
en
u..
00%
w
z:
W%
W%
0
-I
c
(_)
m
<(
"'mm:::c
z:
en
<(
w
-I
0
-"'
;;c
--"""
......
0
~· ~· ~·
s· s'
<--...'
10CS
~· ~· ~·
s·
s'
~~~ ~?~ ~~ # ~~&~.;f # ~~ # ~0~ ~~--\# ~~<v.$- <v~~ .f>~$-#'
~~~'q ~s~~
~
~
~
~
~~r:::i~
~· ~·
~· ~· ~· ~·
s· s·
~· ~·
s
~~~~~~~~~$~~~~~~~~~~~~~~~~
~~~~~~
§>9 ~~~9~~~~~~~&
#~fr~##~
-£?;. ~.::55
~~s
~~
~~OJ
~
s·
P##~&~~~~~~~~~~~~~~~~~~~~~
o
~
~· ~·
~&#~#s##~~#~##~~~g~~~###~~~
q~0~~~~~~~~~~~-~~~~~~8~~~~~
~
c;::.
"""
N
N
N
0%. s'
w
a>
a-,
10%
~
~
c
r
m
......
0Ut
~%
:::::>
-I
N
0
B. AMYLOLIQUEFACIENS (n=32)
00%
en
OJ
~
ro%
I-
"'c
"'=i
GENUS (n=81) ~ B. SUBTILIS GROUP (n=53)
[SIB. AMYLOLIQUEFACIENS SUBSP. PLANTARUM (n=28)
~
c,}~
@
<:/..~~
~
~
,-
~~$S
&
#''V
~
$
#
~--\
~~s4;:-~.P
~
#
<v'5
SUBSYSTEM FEATURES
c}~~$'~s-#'0~~ #s ~~
ef'#
&~
0~ ~
~<§~
~
~'5
~
~
""d
(""l
g
00
N
0
......
FIG. 2B
Ut
0Ut
~
N
~
l.O
Figure 2(~
:.;;
0
N
0
D
a-,
"""
0Ut
""'
N
N
N
fm·
l'IY.".l:A"otbetical Proteins
y;
fm ~rransportatinn
fm
Second:arv•·' J\1et:aboUtes
(:arbon usage
fm I~egub1tion
WI
. '"
~
_..
0
~iJ.11·H')lh~u
:.;.;,~~u·uh.ns
()the rs
""d
(""l
g
00
N
0
"""
Ut
0Ut
~
N
~
l.O
Pigure 3
:.;;
0
N
0
a-,
"""
0Ut
Pseudomonas syringae
pv,. tabaci
""'
N
N
N
Rhizobium radiobader
Xanthomonas campestris
pv~
vesu::at ona
+.
.~
""d
(""l
g
00
N
0
"""
Ut
0Ut
~
N
~
l.O
WO 2016/054222
PCT/US2015/053239
7/10
~
w
t\l•
~'")·
,.,,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..... ,,..
fl'i
~1
({~
0)
....
~-~
·~
~
N
t~)
t~")
\~
WO 2016/054222
PCT/US2015/053239
8/10
CQ
~
QJ
;...
·-=
CJ)
~
WO 2016/054222
9/10
111•
PCT/US2015/053239
WO 2016/054222
PCT/US2015/053239
10/10
~
t'"'
-=
.~
~
~
~
~
'
"
~.
~
"
<
~
~
~
~
-
·~~
~
.~
·'.~..:.-:
~
.....
::r;
=
,.,....
~
•'~
'
~
.~.·
~
;...
~
'~
~
~
"
~
~·
~
~
~
~
~
=
z
••
t~
~''·
~'
!J!!
·"""-
~
~
'"''-
"'~
~<
~
'*'"'
~.
•
...... ·
=
~
............~
I
\C
~
;..,:
·-=
~
~
··«(
~'
~
~:(,,
•
~
,.
®)
_,
4
'"""
M
~
,.__;.;...
~
~~
~
[ -i
INTERNATIONAL SEARCH REPORT
International application No
PCT/US2015/053239
A. CLASSIFICATION OF SUBJECT MATTER
INV.
ADD.
A01N63/00
A61K35/74
According to International Patent Classification (IPC) or to both national classification and IPC
B. FIELDS SEARCHED
Minimum documentation searched (classification system followed by classification symbols)
AOlN A23L A61K
Documentation searched other than minimum documentation to the extent that such documents are included in the fields searched
Electronic data base consulted during the international search (name of data base and, where practicable, search terms used)
EPO-Internal, BIOS IS, EMBASE, FSTA, IBM-TDB
C. DOCUMENTS CONSIDERED TO BE RELEVANT
Category•
x
Citation of document, with indication, where appropriate, of the relevant passages
us 7 422 737 Bl (NUSS I NOV ITCH AMOS [IL] ET
AL) 9 September 2008 (2008-09-09)
claims 1-27
x
Relevant to claim No.
1,2,4-9,
11,12,
15-17,
19-23
- --- -
us 6 280 719 Bl (SUH HYUNG-WON
1,4-9,
11,12,
16,19-23
[KR])
28 August 2001 (2001-08-28)
abstract; claims 2, 8, 16
column 2, 1i ne 48 - 1i ne 51
- --- -
[]]
Further documents are listed in the continuation of Box C.
• Special categories of cited documents :
"A" document defining the general state of the art which is not considered
to be of particular relevance
"E" earlier application or patent but published on or after the international
filing date
"L" document which may throw doubts on priority claim(s) or which is
cited to establish the publication date of another citation or other
special reason (as spec~ied)
"O" document referring to an oral disclosure, use, exhibition or other
means
"P" document published prior to the international filing date but later than
the priority date claimed
Date of the actual completion of the international search
17 December 2015
2
Name and mailing address of the ISA/
European Patent Office, P.B. 5818 Patentlaan 2
NL - 2280 HV Rijswijk
Tel. (+31-70) 340-2040,
Fax: (+31-70) 340-3016
-/--
lRJ
See patent family annex.
"T" later document published after the international filing date or priority
date and not in conflict with the application but cited to understand
the principle or theory underlying the invention
"X" document of particular relevance; the claimed invention cannot be
considered novel or cannot be considered to involve an inventive
step when the document is taken alone
"Y" document of particular relevance; the claimed invention cannot be
considered to involve an inventive step when the document is
combined with one or more other such documents, such combination
being obvious to a person skilled in the art
"&" document member of the same patent family
Date of mailing of the international search report
13/01/2016
Authorized officer
Dumont, Elisabeth
Form PCT/ISA/21 O (second sheet) (April 2005)
page 1 of 3
INTERNATIONAL SEARCH REPORT
International application No
PCT/US2015/053239
C(Continuation).
Category"
X
DOCUMENTS CONSIDERED TO BE RELEVANT
Citation of document, with indication, where appropriate, of the relevant passages
I. V. YEGORENKOVA ET AL: "Composition and
immunochemical characteristics of
exopolysaccharides from the rhizobacterium
Paenibacillus polymyxa 1465",
MICROBIOLOGY,
vol . 77, no. 5,
1 October 2008 (2008-10-01), pages
553-558, XP055236789,
Relevant to claim No.
1
us
ISSN: 0026-2617, DOI:
10.1134/S002626170805007X
abstract
x
HE PENGFEI ET AL: "Genome sequence of the
plant growth promoting strainBacillus
amyloliquefacienssubsp.plantarumB9601-Y2
and expression of mersacidin and other
secondary metabolites",
JOURNAL OF BIOTECHNOLOGY, ELSEVIER SCIENCE
PUBLISHERS, AMSTERDAM, NL,
vol. 164, no. 2,
26 January 2013 (2013-01-26), pages
281-291, XP028990883,
ISSN: 0168-1656, DOI:
10.1016/J.JBIOTEC.2012.12.014
the whole document
in particular abstract; page 283, col. 1,
paragraph 2; Fig. 5; conclusion.
1-34
x
ADNAN NI AZ I ET AL: "Genome Analysis of
Bacillus amyloliquefaciens Subsp.
plantarum UCMB5113: A Rhizobacterium That
Improves Plant Growth and Stress
Management",
PLOS ONE,
vol. 9, no. 8, 13 August 2014 (2014-08-13)
, page e104651, XP055236819,
DOI: 10.1371/journal.pone.0104651
the whole document
in particular page 12, col. 1 and col. 2
1-34
A
CHAO RAN ET AL: "Identification of
Bacillus Strains for Biological Control of
Catfish Pathogens",
PLOS ONE,
vol . 7, no. 9,
21 September 2012 (2012-09-21), page
e45793, XP055236454,
DOI: 10.1371/journal.pone.0045793
the whole document
1-34
-/--
2
Form PCT/ISA/21 O (continuation of second sheet) (April 2005)
page 2 of 3
INTERNATIONAL SEARCH REPORT
International application No
PCT/US2015/053239
C(Continuation).
Category•
X,P
DOCUMENTS CONSIDERED TO BE RELEVANT
Citation of document, with indication, where appropriate, of the relevant passages
KAI WU ET AL: "Pectin Enhances
Bio-Control Efficacy by Inducing
Colonization and Secretion of Secondary
Metabolites by Bacillus amyloliquefaciens
SQY 162 in the Rhizosphere of Tobacco",
PLOS ONE,
vol. 10, no. 5, 21 May 2015 (2015-05-21),
page e0127418, XP055236810,
DOI: 10.1371/journal.pone.0127418
the whole document
Relevant to claim No.
1-9,
11-23
2
Form PCT/ISA/21 O (continuation of second sheet) (April 2005)
page 3 of 3
INTERNATIONAL SEARCH REPORT
International application No
Information on patent family members
us 7422737
Bl
I
09-09-2008 NONE
us 6280719
Bl
28-08-2001
Patent document
cited in search report
I
Publication
date
AU
AU
CA
EP
ID
JP
JP
us
WO
Form PCT/ISA/21 O (patent family annex) (April 2005)
PCT/US2015/053239
I
Patent family
member(s)
735602
5882198
2279489
1015555
22852
3428658
2000513582
6280719
9835017
B2
A
Al
Al
A
B2
A
Bl
Al
Publication
date
12-07-2001
26-08-1998
13-08-1998
05-07-2000
09-12-1999
22-07-2003
17-10-2000
28-08-2001
13-08-1998