BIOINFORMATICS APPLICATIONS NOTE
Vol. 21 no. 9 2005, pages 2136–2137
doi:10.1093/bioinformatics/bti308
Systems biology
Time accelerated Monte Carlo simulations of biological
networks using the binomial τ -leap method
Abhijit Chatterjee, Kapil Mayawala, Jeremy S. Edwards and Dionisios G. Vlachos∗
Department of Chemical Engineering, University of Delaware, Newark, DE 19716, USA
Received on January 9, 2005; revised on February 2, 2005; accepted on February 3, 2005
Advance Access publication February 12, 2005
ABSTRACT
Summary: Developing a quantitative understanding of intracellular
networks requires simulations and computational analyses. However,
traditional differential equation modeling tools are often inadequate
due to the stochasticity of intracellular reaction networks that can
potentially influence the phenotypic characteristics. Unfortunately,
stochastic simulations are computationally too intense for most biological systems. Herein, we have utilized the recently developed
binomial τ -leap method to carry out stochastic simulations of the
epidermal growth factor receptor induced mitogen activated protein
kinase cascade. Results indicate that the binomial τ -leap method
is computationally 100–1000 times more efficient than the exact
stochastic simulation algorithm of Gillespie. Furthermore, the binomial
τ -leap method avoids negative populations and accurately captures
the species populations along with their fluctuations despite the large
difference in their size.
Availability:
http://www.dion.che.udel.edu/multiscale/Introduction.
html. Fortran 90 code available for academic use by email.
Contact: vlachos@che.udel.edu
Supplementary information: Details about the binomial τ -leap
algorithm, software and a manual are available at the above website.
INTRODUCTION
The importance of stochasticity in biological systems is well established by several theoretical studies (Arkin et al., 1998; Morton-Firth
and Bray, 1998; Resat et al., 2003; Meng et al., 2004) and experimental work (Elowitz et al., 2002; Blake et al., 2003). Stochasticity in chemical systems has been extensively studied using the
exact stochastic simulation algorithm (SSA) of Gillespie (1976).
However, SSA is limited to non-stiff systems on short times.
Biological networks exhibit multiple time scales, ranging from
microseconds to days (Edwards and Palsson, 2000), and demand
efficient stochastic simulation algorithms (Lok, 2004). Such recently
developed algorithms are reviewed in Turner et al. (2004) and
Vlachos (2005). The family of τ -leap methods is the only coarsegrained stochastic simulation method that can provide accurate
simulation results.
Here the binomial τ -leap method (Chatterjee et al., 2005), a variant of the original Poisson τ -leap method of Gillespie (2001), is
employed to study the signaling pathway of EGF receptor (EGFR)
activated mitogen activated protein (MAP) kinase cascade. Activated
EGF receptors trigger a rich network of signaling pathways and
∗ To
whom correspondence should be addressed.
2136
regulate cell functions such as proliferation, differentiation and
migration (Yarden and Gur, 2004). In this work, we have used a
mathematical model of this signaling pathway to study the activation of the MAP kinase cascade from surface and internalized EGF
receptors (Schoeberl et al., 2002). The published model is based on
ordinary differential equations (ODEs) of 94 signaling species with
296 reactions (referred to as events in this paper) in a well-mixed
environment. The complexity of the model renders the MAP kinase
cascade an excellent benchmark problem for testing the applicability
of the binomial τ -leap method for biological systems.
THE BINOMIAL τ -LEAP METHOD
The binomial τ -leap method follows the stochastic evolution of
N species among M reactions as an approximate Markov process.
The algorithm requires the initial population size of all species (i.e.
the number of molecules) and the reaction kinetics as inputs. This
information is used to compute transition probabilities per unit time
of all events at time t. A ‘bundle’ of events, sampled from a binomial distribution, are allowed to trigger in a time interval of size τ ,
and the time is incremented to t + τ . This process is repeated as
the algorithm leaps along the time axis. The maximum τ is chosen
to prevent substantial changes in populations (Gillespie, 2001). This
is automatically taken care of using a simple, computationally inexpensive criterion (Chatterjee et al., 2005) according to which the user
can control the speed-up by inputting the coarse-graining factor r of
time stepping in comparison to the SSA. This criterion is chosen
here because r is constrained by mass conservation, 0 < r ≤ 1,
but future work is needed to develop adaptive time step methods.
Previous experience indicates that r up to 0.2 provides accurate
simulations that are much faster than the SSA. A similar binomial τ leap method has been developed independently by Tian and Burrage
(2004).
The algorithm conserves mass at both the single reaction and
the entire network levels by constraining the number of reaction
firings, thereby eliminating for the first time negative populations
in τ -leaping (Chatterjee et al., 2005). Numerical simulations using
simple reaction networks (Chatterjee et al., 2005) demonstrated that
the method is more accurate than the original Poisson τ -leap method
of Gillespie (2001).
RESULTS
Simulations spanning 15 min of real time were performed for
the MAP kinase cascade using the binomial τ -leap method,
© The Author 2005. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oupjournals.org
Time accelerated MC simulations of biological
15
(a)
Species
(a)
SSA 3.0±1.8
r = 0.2 3.0±1.8
r = 0.5 3.0±1.8
1000
10
0.2
5
500
0
5
10
0
15
20
150
0
x 10
5
10
15
4
(b)
(c)
(b)
(c)
59.7±7.5 15224±44
59.0±7.6 15221±48
59.2±7.7 15201±50
Probability
1500
0
0.1
15
100
XMEK-PP
XRaF*
0.3
x 106
2000
XERK-PP
X(EGF-EGF)2
2500
50
0
0
5
10
Time (min)
15
10
0.0
5
0
0
5
10
Time (min)
15
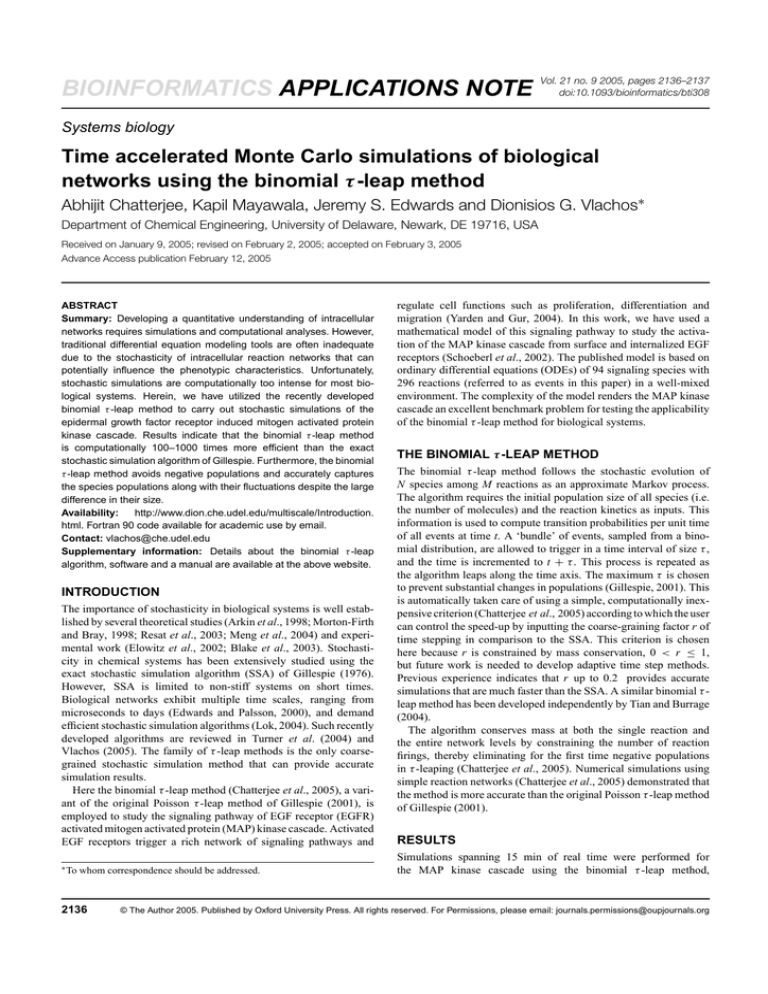
Fig. 1. Time-dependent populations, denoted as Xi (number of molecules)
for species i, using the stochastic simulation algorithm (SSA; thick solid
line) and the binomial τ -leap method (thin solid line, r = 0.2 and squares,
r = 0.5).
with different levels of coarse-graining r = 0.1–0.5, and
the SSA. Reaction dynamics and initial populations are specified on the Website (http://www.dion.che.udel.edu/multiscale/
Introduction.html). Intracellular populations, namely free dimerized
EGF bound EGFR ((EGF-EGFR)2), doubly phosphorylated ERK
(ERK-PP), doubly phosphorylated MEK (MEK-PP) and free and
activated RaF (RaF*), are plotted against time in Figure 1 for selected species whose populations span from small to large ones. The
binomial τ -leap method accurately captures the network kinetics for
all times despite the large disparity in species population size in this
complex network. The Poisson τ -leap method on the other hand
encounters negative populations since small populations are present
(Gillespie, 2001).
Figure 2 compares fluctuations of the two methods using the probability density functions obtained at time t = 1 min (with ensemble
averages over 500 trajectories for the SSA and 1000 for the binomial
τ -leap method). The mean and standard deviation for representative
species, namely internalized dimerized EGF bound EGFR ((EGFEGFRi)2), internalized EGFR (EGFRi) and activated (EGF-EGFR)2
(EGF-EGFR*2), that span many orders of magnitude in number, are
tabulated in the inset of Figure 2. The consistency between the methods for the mean, standard deviation and higher order moments is
indicative of the accuracy that can be obtained using the binomial
τ -leap method.
The SSA required 3 days on a DEC alpha 833 MHz processor
for the simulation depicted in Figure 1 as compared to 6–30 min
taken using the τ -leap method. The binomial τ -leap is 100–1000
times more efficient than the SSA over the range of r values
explored. The low computational requirements of the binomial τ leap method enable accurate long time simulations that are beyond
the reach of SSA (see example on the Website). Due to its accuracy,
robustness, simplicity and substantial speed-up, the binomial τ leap method appears to be a particularly promising, approximate
stochastic simulation method for computational biology.
0
10 30
X(EGF-EGFRi)2
XEGFRi
90
15400
15000
X(EGF-EGFR*)2
Fig. 2. Probability density functions (pdf) using the stochastic simulation
algorithm (SSA, circles) and the binomial τ -leap method at time t = 1
min (squares, r = 0.2; diamonds, r = 0.5). Populations for species i
are denoted as Xi (number of molecules). The solid lines are curve fits for
visual aid. Average populations and standard deviation are tabulated in the
inset.
ACKNOWLEDGEMENTS
This work was partially supported by the NSF through CTS-0312117.
REFERENCES
Arkin,A.P. et al. (1998) Stochastic kinetic analysis of developmental pathway bifurcation in phage l-infected Escherichia coli cells. Genetics, 149,
1633–1648.
Blake,W.J. et al. (2003) Noise in eukaryotic gene expression. Nature, 422, 633–637.
Chatterjee,A. et al. (2005) Binomial distribution based τ -leap accelerated stochastic
simulation. J. Chem. Phys., 122, 024112.
Edwards,J.S. and Palsson,B.O. (2000) Multiple steady states in kinetic models of red
cell metabolism. J. Theoret. Biol., 207, 125–127.
Elowitz,M.B. et al. (2002) Stochastic gene expression in a single cell. Science, 297,
1183–1186.
Gillespie,D.T. (1976) A general method for numerically simulating the stochastic
evolution of coupled chemical reactions. J. Comput. Phys., 22, 403–434.
Gillespie,D.T. (2001) Approximate accelerated stochastic simulation of chemically
reacting systems. J. Chem. Phys., 115, 1716–1733.
Lok,L. (2004) The need for speed in stochastic simulation. Nat. Biotechnol., 22,
964–965.
Meng,T.C. et al. (2004) Modeling and simulation of biological systems with stochasticity. In Silico Biol., 4, 0024.
Morton-Firth,C.J. and Bray,D. (1998) Predicting temporal fluctuations in an intracellular
signalling pathway. J. Theoret. Biol., 192, 117–128.
Resat,H. et al. (2003) An integrated model of epidermal growth factor receptor trafficking
and signal transduction. Biophys. J., 85, 730–743.
Schoeberl,B. et al. (2002) Computational modeling of the dynamics of the MAP
kinase cascade activated by surface and internalized receptors. Nat. Biotechnol., 20,
370–375.
Tian,T. and Burrage,K. (2004) Binomial leap methods for simulating stochastic chemical
kinetics. J. Chem. Phys., 121, 10356–10364.
Turner,T.E. et al. (2004) Stochastic approaches for modelling in vivo reactions. Comput.
Biol. Chem., 28, 165–178.
Vlachos,D.G. (2005) The emerging field of multiscale analysis: a review with examples
from systems biology, materials engineering, and fluid-surface interacting systems.
Adv. Chem. Eng, in press.
Yarden,Y. and Gur,G. (2004) Enlightened receptor dynamics. Nat. Biotechnol., 22,
169–170.
2137