Parallel Computing 32 (2006) 222–230
www.elsevier.com/locate/parco
Pipeline Givens sequences for computing the QR
decomposition on a EREW PRAM q
Marc Hofmann
a
a,*
, Erricos John Kontoghiorghes
b,c
Institut d’informatique, Université de Neuchâtel, Emile-Argand 11, Case Postale 2, CH-2007 Neuchâtel, Switzerland
b
Department of Public and Business Administration, University of Cyprus, Cyprus
c
School of Computer Science and Information Systems, Birkbeck College, University of London, UK
Received 4 March 2005; received in revised form 25 September 2005; accepted 19 November 2005
Available online 18 January 2006
Abstract
Parallel Givens sequences for computing the QR decomposition of an m · n (m > n) matrix are considered. The Givens
rotations operate on adjacent planes. A pipeline strategy for updating the pair of elements in the affected rows of the
matrix is employed. This allows a Givens rotation to use rows that have been partially updated by previous rotations.
Two new Givens schemes, based on this pipeline approach, and requiring respectively n2/2 and n processors, are developed.
Within this context a performance analysis on an exclusive-read, exclusive-write (EREW) parallel random access machine
(PRAM) computational model establishes that the proposed schemes are twice as efficient as existing Givens sequences.
2005 Elsevier B.V. All rights reserved.
Keywords: Givens rotation; QR decomposition; Parallel algorithms; PRAM
1. Introduction
Consider the QR decomposition of the full column rank matrix A 2 Rmn :
R n
T
;
Q A¼
0 mn
ð1Þ
where Q 2 Rmm and R is upper triangular of order n. The triangular matrix R in (1) is derived iteratively from
e iþ1 , where A
e 0 ¼ A and Qi is orthogonal: the triangularization process terminates when A
e v (v > 0) is
ei ¼ A
QTi A
upper triangular. Q = Q0 Q1 Qv is not computed explicitly. R can be derived by employing a sequence of
Givens rotations. The Givens rotation (GR) that annihilates the element Ai,j when applied from the left of
A has the form
q
*
This work is in part supported by the Swiss National Foundation Grants 101412-105978.
Corresponding author. Fax: +41 32 718 27 01.
E-mail addresses: marc.hofmann@unine.ch (M. Hofmann), erricos@ucy.ac.cy (E.J. Kontoghiorghes).
0167-8191/$ - see front matter 2005 Elsevier B.V. All rights reserved.
doi:10.1016/j.parco.2005.11.001
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
e i;j ; I mi Þ
Gi;j ¼ diag ðI i2 ; G
e i;j ¼
with G
c
s
s
c
;
where c = Ai1,j/t, s = Ai,j/t and t2 ¼ A2i1;j þ A2i;j ð6¼ 0Þ.
(a) SK
(b) mSK
Fig. 1. The SK and modified SK Givens sequences to compute the QR decomposition, where m = 16 and n = 6.
Fig. 2. Partial annihilation of a matrix by the PipSK scheme when n = 6.
223
224
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
A GR affects only the ith and (i 1)th rows of A. Thus, bm/2c rotations can be applied simultaneously. A
compound disjoint Givens rotation (CDGR) comprises rotations that can be applied simultaneously. Parallel
algorithms for computing the QR decomposition based on CDGRs have been developed [2,3,8–10,12–14]. The
Greedy sequence in [4,11] was found to be optimal; that is, it requires less CDGRs than any other Givens
strategy. However, the computation of the indices of the rotations which do not involve adjacent planes is
non-trivial. The employment of rotations between adjacent planes facilitates the development of efficient factorization strategies for structured matrices [6].
An EREW (exclusive-read, exclusive-write) PRAM (parallel random access machine) computational model
is considered [6]. It is assumed that there are p processors which can perform simultaneously p GRs. A single
time unit is defined to be the time required to execute the operation of applying a Givens rotation to two oneelement vectors. Thus, the elapsed time necessary to perform a rotation depends on the length of the vectors
involved. Computing c and s requires 6 flops. Rotating two elements requires another six flops. Hence, annihilation of an element and performing the necessary updating of an m · n matrix requires 6n flops. Notice that
the GR is not applied to the first pair of elements the components of which are set to t and zero, respectively.
To simplify the complexity analysis of the proposed algorithms it is assumed that m and n are even. Complexities are given in time units.
(a) PGS-1
(b) PGS-2
(c) PGS-2 cycles
Fig. 3. Parallel Givens sequences for computing the QR decomposition, where m = 16 and n = 6.
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
225
New parallel Givens sequences to compute the QR decomposition are proposed. Throughout the execution
of a Givens sequence the annihilated elements are preserved. In the next section a pipelined version of the parallel Sameh and Kuck scheme is presented. Two new pipeline parallel strategies are discussed in Section 3. A
theoretical analysis of complexities of the various schemes is presented. Section 4 summarizes what has been
achieved.
2. Pipeline parallel SK sequence
The parallel Sameh and Kuck (SK) scheme in [14] computes (1) by applying up to n GRs simultaneously.
Each GR is performed by one processor. Specifically, the elements of the ith column of A begin to be annihilated by the (2i 1)th CDGR as illustrated in Fig. 1(a). The numeral i and the symbol • denote an element
annihilated by the ith CDGR and a non-zero element, respectively. The number of CDGRs and time units
required by the SK scheme are given, respectively, by
C SK ðm; nÞ ¼ m þ n 2
Fig. 4. Triangularization of a 16 · 6 matrix by the PipPGS-1 algorithm.
226
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
and
T SK ðm; nÞ ¼ ðm 1Þn þ
n
X
ðn j þ 1Þ ¼ nð2m þ n 3Þ=2.
j¼2
Here it is assumed that p = n.
ðkÞ
Let gi;j (k = 1, . . . , n j + 1) denote the application of Gi,j to the kth pair of elements in positions
(i, j + k 1) and (i 1, j + k 1). That is, the rotation Gi,j operating on the (n j + 1)—element subvectors
ð1Þ
ðnjþ1Þ
of rows i and i 1 is now expressed as the application of a sequence of elementary rotations gi;j ; . . . ; gi;j
to
the pairs of elements {(i, j), (i 1, j)}, . . . , {(i, n), (i 1, n)}, respectively. The rotation Gi+1,j+1 can begin before
ð1Þ
ð2Þ
the application of Gi,j has been completed. Specifically, giþ1;jþ1 can be applied once gi;j has been executed.
Thus, several GRs can operate concurrently on different elements of a row. The Pipelined SK (PipSK) scheme
employs this strategy. The first steps of the PipSK scheme are illustrated in Fig. 2. As in the case of the SK
scheme, PipSK initiates a CDGR every second time unit and its overall execution time is given by
T PipSK ðm; nÞ ¼ 2C SK ðm; nÞ 1 ¼ 2m þ 2n 5.
The number of CDGRs being performed
is n/2 requiring 2i processors each (i = 1, . . . , n/2). The
Pn=2 in parallel
PipSK scheme thus requires p ¼ i¼1
2i n2 =4 processors. Its annihilation pattern—a modified SK
scheme—is shown in Fig. 1(b).
Fig. 5. Annihilation cycle of the PipPGS-1 algorithm when n = 6.
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
Fig. 6. Annihilation cycle of the PGS-2 scheme when n = 6.
Fig. 7. Annihilation cycle of the PipPGS-2 algorithm when n = 6.
227
228
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
3. New pipeline parallel Givens sequences
Alternative parallel Givens (PG) sequences to compute the QR decomposition and which are more suitable
for pipelining are shown in Fig. 3. The number of CDGRs applied by the first PG sequence (PGS-1 [1]) is
given by
C PGS-1 ðm; nÞ ¼ m þ 2n 3.
The Pipelined PGS-1 (PipPGS-1) is illustrated in Fig. 4. It requires p n2/2 processors. It initiates a CDGR
every time unit and its time complexity is given by the total number of CDGRs applied. That is,
T PipPGS-1 ðm; nÞ ¼ C PGS1 ðm; nÞ ¼ m þ 2n 3.
The PipPGS-1 operates in cycles of n + 1 time units. This is shown in Fig. 5. In every time unit up to n
processors initiate a GR, while the other processors are updating previously initiated rotations. Each processor executes two GRs in one cycle with complexities T1 and T2 respectively, such that T1 + T2 = n + 1 time
units. The PipPGS-1 performs better than the SK scheme utilizing approximately n/2 times the number of processors. That is, TSK(m, n)/TPipPGS-1(m, n) n. Hence, the efficiency of the PipPGS-1 is twice that of the SK
scheme.
The PGS-2 illustrated in Fig. 3(b) employs p = n processors. A cycle involves one CDGR and n consecutive
GRs and annihilates up to 2n elements in n + 1 steps. This is illustrated in Fig. 6. The PGS-2 scheme requires
more steps than the SK scheme. When m and n are even, the sequence consists of (m + n 2)/2 cycles which
Fig. 8. Initial annihilation steps of a 16 · 6 matrix by the PipPGS-2 algorithm.
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
229
Fig. 9. Final annihilation steps of a 16 · 6 matrix by the PipPGS-2 algorithm.
can be partitioned in three sets comprising cycles {1, . . . , n 1}, {n, . . . , (m 2)/2} and {m/2, . . . , (m + n 2)/
2}. The sets, cycles and constituent steps are detailed in Fig. 3. The ith cycle in the first, second and third set
applies i + 1, n + 1 and 2(n/2 i + 1) steps, respectively. The total number of steps applied by the PGS-2 is
given by
C PGS-2 ðm; nÞ ¼
n1
X
ði þ 1Þ þ ðm 2nÞðn þ 1Þ=2 þ
n=2
X
2i ð2mn þ 2m n2 Þ=4.
i¼1
i¼1
The Pipelined PGS-2 (PipPGS-2) applies 2n GRs in a pipeline manner. Fig. 7 illustrates the annihilation
cycle of the PipPGS-2 which annihilates 2n elements in n + 1 time units. This is equivalent to two CDGRs
of the SK scheme for which 2n time units are required. Figs. 8 and 9 illustrate, respectively, the initial and
the final phase of the annihilation process of a 16 · 6 matrix by the PipPGS-2. An asterisk denotes the start
of a new cycle.
In every time unit a processor initiates a GR. When m and n are even, the first (m 2)/2 cycles are executed
in n + 1 time units each. The ith cycle in the third set requires 2(n/2 i + 1) + 1 time units (i = 1, . . . , n/2). The
overall execution time of the PipPGS-2 algorithm is given by
T PipPGS-2 ðm; nÞ ¼ ðm 2Þðn þ 1Þ=2 þ
n=2
X
ð2i þ 1Þ ð2mn þ 2m þ n2 Þ=4.
i¼1
Furthermore, if m n and for the same number of processors,
T SK ðm; nÞ
2.
T PipPGS-2 ðm; nÞ
That is, the proposed scheme is twice as fast as the SK scheme.
230
M. Hofmann, E.J. Kontoghiorghes / Parallel Computing 32 (2006) 222–230
Table 1
Summary of the complexities of the SK and pipelined schemes
Scheme
Processors
Complexity
SK
n
n(2m + n 3)/2
PipSK
PipPGS-1
PipPGS-2
2
n /4
n2/2
n
2m + 2n 5
m + 2n 3
(2mn + 2m + n2)/4
4. Conclusions
A new pipeline parallel strategy has been proposed for computing the QR decomposition. Its computational complexity is compared to the SK scheme of Sameh and Kuck in [14]. The complexity analysis is
not based on the unrealistic assumption that all CDGRs, or cycles, have the same execution time. Instead,
the number of operations performed by a single Givens rotation is given by the size of the pair of vectors used
in the rotation. It was found that for an equal number of processors the pipelined scheme solves the m · n QR
decomposition twice as fast as the SK scheme when m n. The complexities are summarized in Table 1.
Block versions of the SK scheme have previously been designed to compute the orthogonal factorizations of
structured matrices which arise in econometric estimation problems [5,15]. Within this context the Givens rotations are replaced by orthogonal factorizations which employ Householder reflections. Thus, it might be fruitful to investigate the effectiveness of incorporating the pipeline strategy in the design of block algorithms [7,16].
Acknowledgements
The authors are grateful to Maurice Clint, Ahmed Sameh and the anonymous referee for their valuable
comments and suggestions.
References
[1] A. Bojanczyk, R. Brent, H. Kung, Numerically stable solution of dense systems of linear equations using mesh-connected processors,
SIAM J. Sci. Statist. Comput. 5 (1984) 95–104.
[2] M. Cosnard, M. Daoudi, Optimal algorithms for parallel Givens factorization on a coarse-grained PRAM, J. ACM 41 (2) (1994) 399–
421.
[3] M. Cosnard, J.-M. Muller, Y. Robert, Parallel QR decomposition of a rectangular matrix, Numerische Mathematik 48 (1986) 239–
249.
[4] M. Cosnard, Y. Robert, Complexité de la factorisation QR en parallèle, C. R. Acad. Sci. Paris, ser. I 297 (1983) 137–139.
[5] E.J. Kontoghiorghes, Parallel Algorithms for Linear Models: Numerical Methods and Estimation Problems, Advances in
Computational Economics, volume 15, Kluwer Academic Publishers, Boston, MA, 2000.
[6] E.J. Kontoghiorghes, Parallel Givens sequences for solving the general linear model on a EREW PRAM, Parallel Algorithms Appl.
15 (1–2) (2000) 57–75.
[7] E.J. Kontoghiorghes, Parallel strategies for rank-k updating of the QR decomposition, SIAM J. Matrix Anal. Appl. 22 (3) (2000)
714–725.
[8] E.J. Kontoghiorghes, Greedy Givens algorithms for computing the rank-k updating of the QR decomposition, Parallel Comput. 28
(9) (2002) 1257–1273.
[9] F.T. Luk, A rotation method for computing the QR decomposition, SIAM J. Sci. Statist. Comput. 7 (2) (1986) 452–459.
[10] J.J. Modi, Parallel Algorithms and Matrix Computation, Oxford Applied Mathematics and Computing Science series, Oxford
University Press, 1988.
[11] J.J. Modi, M.R.B. Clarke, An alternative Givens ordering, Numerische Mathematik 43 (1984) 83–90.
[12] A.H. Sameh, Solving the linear least squares problem on a linear array of processors, in: Algorithmically Specialized Parallel
Computers, Academic Press, Inc., 1985, pp. 191–200.
[13] A.H. Sameh, R.P. Brent, On Jacobi and Jacobi like algorithms for a parallel computer, Math. Comput. 25 (115) (1971) 579–590.
[14] A.H. Sameh, D.J. Kuck, On stable parallel linear system solvers, J. ACM 25 (1) (1978) 81–91.
[15] P. Yanev, P. Foschi, E.J. Kontoghiorghes, Algorithms for computing the QR decomposition of a set of matrices with common
columns, Algorithmica 39 (2004) 83–93.
[16] P. Yanev, E.J. Kontoghiorghes, Efficient algorithms for block downdating of least squares solutions, Appl. Numer. Math. 49 (2004)
3–15.
Computational Statistics & Data Analysis 52 (2007) 16 – 29
www.elsevier.com/locate/csda
Efficient algorithms for computing the best subset regression
models for large-scale problems夡
Marc Hofmanna,∗ , Cristian Gatua, d , Erricos John Kontoghiorghesb, c
a Institut d’Informatique, Université de Neuchâtel, Switzerland
b Department of Public and Business Administration, University of Cyprus, Cyprus
c School of Computer Science and Information Systems, Birkbeck College, University of London, UK
d Faculty of Computer Science, “Alexandru Ioan Cuza” University of Iasi, Romania
Available online 24 March 2007
Abstract
Several strategies for computing the best subset regression models are proposed. Some of the algorithms are modified versions of
existing regression-tree methods, while others are new. The first algorithm selects the best subset models within a given size range.
It uses a reduced search space and is found to outperform computationally the existing branch-and-bound algorithm. The properties
and computational aspects of the proposed algorithm are discussed in detail. The second new algorithm preorders the variables
inside the regression tree. A radius is defined in order to measure the distance of a node from the root of the tree. The algorithm
applies the preordering to all nodes which have a smaller distance than a certain radius that is given a priori. An efficient method
of preordering the variables is employed. The experimental results indicate that the algorithm performs best when preordering
is employed on a radius of between one quarter and one third of the number of variables. The algorithm has been applied with
such a radius to tackle large-scale subset-selection problems that are considered to be computationally infeasible by conventional
exhaustive-selection methods. A class of new heuristic strategies is also proposed. The most important of these is one that assigns
a different tolerance value to each subset model size. This strategy with different kind of tolerances is equivalent to all exhaustive
and heuristic subset-selection strategies. In addition the strategy can be used to investigate submodels having noncontiguous size
ranges. Its implementation provides a flexible tool for tackling large scale models.
© 2007 Elsevier B.V. All rights reserved.
Keywords: Best-subset regression; Regression tree; Branch-and-bound algorithm
1. Introduction
The problem of computing the best-subset regression models arises in statistical model selection. Most of the criteria
used to evaluate the subset models rely upon the residual sum of squares (RSS) (Searle, 1971; Sen and Srivastava,
1990). Consider the standard regression model
y = A + ε,
夡 The
∼ (0, 2 Im ),
R routines can be found at URL: http://iiun.unine.ch/matrix/software.
∗ Corresponding author. Tel.: +41 32 7182708; fax: +41 32 7182701.
E-mail addresses: marc.hofmann@unine.ch (M. Hofmann), cristian.gatu@unine.ch (C. Gatu), erricos@ucy.ac.cy (E.J. Kontoghiorghes).
0167-9473/$ - see front matter © 2007 Elsevier B.V. All rights reserved.
doi:10.1016/j.csda.2007.03.017
(1)
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
17
Table 1
Leaps and BBA: execution times in seconds for data sets of different sizes, without and with variable preordering
# Variables
36
37
38
39
40
41
42
43
44
45
46
47
48
Leaps
BBA
8
2
29
5
44
12
30
8
203
35
57
14
108
9
319
55
135
27
316
37
685
97
2697
380
6023
1722
Leaps-1
BBA-1
3
1
16
4
28
13
9
2
82
20
33
11
22
4
203
47
79
18
86
15
306
51
1326
216
1910
529
where y ∈ Rm , A ∈ Rm×n is the exogenous data matrix of full column rank, ∈ Rn is the coefficient vector and ε ∈ Rn
is the noise vector. The columns of A correspond to the exogenous variables V = [v1 , . . . , vn ]. A submodel S of (1)
comprises some of the variables in V. There are 2n − 1 possible subset models, and their computation is only feasible
for small values of n. The dropping column algorithm (DCA) derives all submodels by generating a regression tree
(Clarke, 1981; Gatu and Kontoghiorghes, 2003; Smith and Bremner, 1989). The parallelization of the DCA moderately
improves its practical value (Gatu and Kontoghiorghes, 2003). Various procedures such as the forward, backward and
stepwise selection try to identify a subset by inspecting very few combinations of variables. However, these methods
rarely succeed in finding the best submodel (Hocking, 1976; Seber, 1977). Other approaches for subset-selection include
ridge regression, the nonnegative garrote and the lasso (Breiman, 1995; Fan and Li, 2001; Tibshirani, 1996). Sequential
replacement algorithms are fairly fast and can be used to give some indication of the maximum size of the subsets that
are likely to be of interest (Hastie et al., 2001). The branch-and-bound algorithms for choosing a subset of k features
from a given larger set of size n have also been investigated within the context of feature selection problems (Narendra
and Fukunaga, 1997; Roberts, 1984; Somol et al., 2004). These strategies are used when the size k of the subset to be
selected is known. Thus, they search over n!/(k!(n − k)!) subsets.
A computationally efficient branch-and-bound algorithm (BBA) has been devised (Gatu and Kontoghiorghes, 2006;
Gatu et al., 2007). The BBA avoids the computation of the whole regression tree and it derives the best subset model
for each number of variables. That is, it computes
argmin RSS(S) subject to |S| = k for k = 1, . . . , n.
(2)
S
The BBA was built around the fundamental property
RSS(S1 ) RSS(S2 )
if S1 ⊆ S2 ,
(3)
where S1 and S2 are two variable subsets of V (Gatu and Kontoghiorghes, 2006). The BBA-1, which is an extension of
the BBA, preorders the n variables according to their strength in the root node. The variables i and j are arranged such that
RSS(V−i ) RSS(V−j ) for each i j , where V−i is the set V from which the ith variable has been deleted. The BBA-1
has been shown to outperform the previously introduced leaps-and-bounds algorithm (Furnival and Wilson, 1974).
Table 1 shows the execution times of the BBA and leaps-and-bounds algorithm for data sets with 36–48 variables.
Note that the BBA outperforms the leaps-and-bounds with preordering in the root node (Leaps-1). A heuristic version
of the BBA (HBBA) that uses a tolerance parameter to relax the BBA pruning test has been discussed. The HBBA
might not provide the optimal solution, but the relative residual error (RRE) of the computed solution is smaller than
the tolerance employed.
Often models within a given size range must be investigated. These models, hereafter called subrange subset models,
do not require the generation of the whole tree. Thus, the adaptation of the BBA for deriving the subrange subset models
is expected to have a lower computational cost, and thus, it can be feasible to tackle larger scale models. The structural
properties of a regression tree strategy which generates the subrange subset models is investigated and its theoretical
complexity derived. A new nontrivial preordering strategy that outperforms the BBA-1 is designed and analyzed. The
new strategy, which can be found to be significantly faster than existing ones, can derive the best subset models from
a larger pool of variables. In addition, some new heuristic strategies based on the HBBA are developed. The tolerance
parameter is either a function of the level in the regression tree, or of the size of the subset model. The novel strategies
decrease execution time while selecting models of similar, or of even better, quality.
18
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
The proposed strategies, which outperform the existing subset selection BBA-1 and its heuristic version, are aimed
at tackling large-scale models. The next section briefly discusses the DCA, and it introduces the all-subset-models
regression tree. It generalizes the DCA so as to select only the submodels within a given size range. Section 3 discusses
a novel strategy that preorders the variables of the nodes in various levels of the tree. The significant improvement in
the computational efficiency when compared to the BBA-1 is illustrated. Section 4 presents and compares various new
heuristic strategies. Theoretical and experimental results are presented. Conclusions and proposals for future work are
discussed in Section 5.
The algorithms were implemented in C++ and are available in a package for the R statistical software environment
(R Development Core Team, 2005). The GNU compiler collection was used to generate the shared libraries. The tests
were run on a Pentium-class machine with 512 Mb of RAM in a Linux environment. Real and artificial data have been
used in the experiments. A set of artificial variables has been randomly generated. The response variable of the true
model is based on a linear combination of a subset of these artificial variables with the addition of some noise. An
intercept term is included in the true model.
2. Subrange model selection
The DCA employs a straightforward approach to solve the best-subset problem (2). It enumerates and evaluates all
possible 2n − 1 subsets of V. It generates a regression tree consisting of 2n−1 nodes (Gatu and Kontoghiorghes, 2003;
Smith and Bremner, 1989). Each node in the tree corresponds to a subset S = [s1 , . . . , sns ] of ns variables and to an
index k (k = 0, . . . , ns − 1). The ns − k subleading models [s1 , . . . , sk+1 ], . . . , [s1 , . . . , sns ] are evaluated. A new node
is generated by deleting a variable. The descending nodes are given by
(drop(S, k + 1), k), (drop(S, k + 2), k + 1), . . . , (drop(S, ns − 1), ns − 2).
Here, the operation drop(S, i) computes a new subset which corresponds to the subset S from which the ith variable
has been deleted. This is equivalent to downdating the QR decomposition after the ith column has been deleted (Golub
and Van Loan, 1996; Kontoghiorghes, 2000; Smith and Bremner, 1989). The DCA employs Givens rotations to move
efficiently from one node to another.
The search space of all possible variable subset models can be reduced by imposing bounds on the size of the subset
models. The subrange model selection problem is to derive:
Sj∗ = argmin RSS(S)
S
subject to |S| = j for j = na , . . . , nb ,
(4)
where na and nb are the subrange bounds (1 na nb n). The DCA and Subrange DCA (RangeDCA) are equivalent
when na = 1 and nb = n. The RangeDCA generates a subtree of the original regression tree. The nodes (S, k) are
not computed when ns < na or k nb . This is illustrated in Fig. 1. The DCA regression tree with n = 5 variables is
shown. The blank nodes represent the RangeDCA subtree for na = nb = 3. Portions of the tree that are not computed
by the RangeDCA are shaded. The nodes in the last two levels of the tree evaluate subset models of sizes 1 and 2 , i.e.
Level
L0
12345
•2345
L1
•345
L2
L3
•45
L4
•5
2•45
3•5
2•5
1•345
23•5
1•45
13•5
12•45
12•5
1•5
Fig. 1. The RangeDCA subtree, where n = 5 and na = nb = 3.
123•5
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
19
the subsets [4], [5], [4, 5], [3, 5], [2, 5] and [1, 5]. These nodes are discarded by the RangeDCA (case ns < na ). The
leftmost node in the tree evaluates the subset model [1, 2, 3, 5] of size 4 . The RangeDCA discards this node (case
k nb ). The Appendix provides a detailed and formal analysis of the RangeDCA.
The branch-and-bound strategy can be applied to the subtree generated by the RangeDCA. This strategy is called
RangeBBA and is summarized in Algorithm 1. The RangeBBA stores the generated nodes of the regression subtree in
a list. The list is managed according to a last-in, first-out (LIFO) policy. The RSSs of the best subset models are recorded
in a table r. The entry ri holds the RSS of the best current submodel of size i. The initial residuals table may be given a
priori based on some earlier results, otherwise the initial residuals are set to positive infinity. The entries are sorted in
decreasing order. Each iteration removes a node (S, k) from the list. The subleading model [s1 , . . . , si ] is evaluated and
compared to the entry ri in the residuals table for i = k + 1, . . . , ns . The entry ri is updated when RSS([s1 , . . . , si ]) < ri .
If ns na , then no child nodes are computed and the iteration terminates; otherwise, the cutting test bS > ri is computed
for i = k + 1, . . . , min(nb , ns − 1) and bS = RSS(S). If the test fails the child node (drop(S, i), i − 1) is generated and
inserted into the node list. Note that, if i < na , then the value rna is used in the cutting test. This is illustrated on Line 11
of the algorithm. The modified cutting test is more efficient than that of the BBA, since rna ri for i = 1, . . . , na − 1.
The algorithm terminates when the node list is empty. Notice that the RangeBBA with preordering (RangeBBA-1) is
obtained by sorting the variables in the initial set V. The RangeBBA outperforms the standard BBA, since it uses a
reduced search space and a more efficient cutting test.
Algorithm 1. The subrange BBA (RangeBBA)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
procedure RangeBBA(V , na , nb , r)
insert (V , 0) into the node list
while the node list is not empty do
Extract (S, k) from the node list
ns ← |S|
Update the residuals rk+1 , . . . , rns
if ns > na then
bS ← RSS(S)
for i = k + 1, . . . , min(nb , ns − 1) do
j ← max(i, na )
if bS > rj go to line 3
S ← drop(S, i)
Insert (S , i − 1) into the node list
end for
end if
end while
end procedure
The effects of the subrange bounds na and nb on the computational performance of the RangeDCA and RangeBBA-1
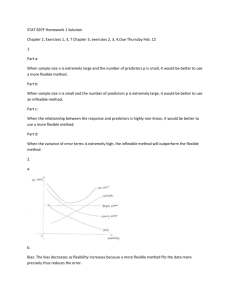
have been investigated. The Fig. 2(a) and (b) show the execution times of the RangeDCA for n = 20 variables and
RangeBBA-1 for 36 variables, respectively. It can be observed that the RangeDCA is computationally effective in
two cases: for narrow size ranges (i.e. nb − na < 2) or extreme ranges (i.e. na = 1 and nb < n/4 or na > 3n/4
and nb = n). The RangeBBA-1, on the other hand, is effective for all ranges such that na > n/2. This is further
confirmed by the results in Table 2. The number of nodes generated by the RangeBBA-1 for the 15 variable POLLUTE data set (Miller, 2002) is shown. All possible subranges 1na nb 15 are considered. For the case na = 1
and nb = 15 the RangeBBA-1 generates 381 nodes and is equivalent to the BBA-1 (Gatu and Kontoghiorghes,
2006).
3. Radius preordering
The BBA with an initial preordering of the variables in the root node (BBA-1) significantly increases the computational speed. The cost of preordering the variables once is negligible. The aim is to consider a strategy that applies
the preordering of variable subsets inside the regression tree and that yields a better computational performance than
20
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
0.8
1.0
0.6
time
time
0.4
0.5
0.2
0.0
5
n_a
0.0
20
n_b
15
10
15
(a)
n_a
10
10
5
20
10
30
20
RangeDCA (n = 20)
30
n_b
20
(b) RangeBBA-1 (n = 36)
Fig. 2. Subrange model selection: execution times in seconds for varying na and nb .
Table 2
Number of nodes generated by the RangeBBA-1 to compute the best subset models of the POLLUTE data set for different size ranges
na
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
nb
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
12
37
31
96
90
75
178
172
157
123
276
270
255
221
173
332
326
311
277
229
103
356
350
335
301
253
127
52
373
367
352
318
270
144
69
38
375
369
354
320
272
146
71
40
11
376
370
355
321
273
147
72
41
12
11
377
371
356
322
274
148
73
42
13
12
12
378
372
357
323
275
149
74
43
14
13
13
13
380
374
359
325
277
151
76
45
16
15
15
15
15
381
375
360
326
278
152
77
46
17
16
16
16
16
15
381
375
360
326
278
152
77
46
17
16
16
16
16
15
1
the BBA-1. The new strategy is hereafter called radius preordering BBA (RadiusBBA). The RadiusBBA sorts the
variables according to their strength. The strength of the ith variable is given by its bound RSS(S−i ) = RSS(drop(S, i)).
The main tool for deriving the bound is the downdating of the QR decomposition after the corresponding column of the data matrix has been deleted. This has a cost, and therefore care must be taken to apply a preordering to the nodes whenever the expected gain outweighs the total cost inherent to the preordering
process.
The RadiusBBA preorders the variables at the roots of large subtrees. The size of the subtree with root (S, k) is
given by 2d−1 , where ns is the number of variables in S and d = ns − k is the number of active variables. The
RadiusBBA defines the node radius = n − d, where n is the initial number of variables in the root node (V , 0). The
radius of a node is a measure of its distance from the root node. Notice that the root nodes of larger subtrees have
a smaller radius, while the roots of subtrees with the same number of nodes have the same radius. Given a radius P
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
21
12345
ρ=0
2345
ρ=1
•345
ρ=2
•45
ρ=3
2•45
ρ=3
3•5
ρ=4
1•345
ρ=2
23•5
ρ=4
2•5
ρ=4
1•45
ρ=3
12•45
ρ=3
13•5
ρ=4
123•5
ρ=4
12•5
ρ=4
1•5
ρ=4
•5
ρ=4
Fig. 3. Nodes that apply preordering for radius P = 3.
the preordering of variables is applied only to nodes of radius < P , where 0 P n. If P = 0 and P = 1, then the
RadiusBBA is equivalent to the BBA and BBA-1, respectively. If P = n, then the active variables are preordered in all
nodes. Fig. 3 illustrates the radius of every node in a regression tree for n = 5 variables. Shaded nodes are preordered
by the RadiusBBA, where P = 3.
The RadiusBBA is illustrated in Algorithm 2. A node is extracted from the node list at each iteration. If the node radius
is less than the given preordering radius P, then the active variables are preordered before updating the residuals table.
The nodes which cannot improve the current best solutions are not generated. The cutting test (see line 11) compares
the bound of the current node to the corresponding entry in the residuals table in order to decide whether or not to
generate a child node.
Algorithm 2. The radius brnach-and-bound algorithm (RadiusBBA)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
procedure RadiusBBA(V , P , r)
n ← |V |
Insert (V , 0) into node list
while node list is not empty do
Extract (S, k) from nodes list
ns ← |S|; ← n − k
if < P then preorder [sk+1 , . . . , sns ]
Update residuals rk+1 , . . . , rns
bS ← RSS(S)
for i = k + 1, . . . , ns − 1 do
if bS > ri goto line 3
S ← drop(S, i)
Insert (S , i − 1) into node list
end for
end while
end procedure
The preordering process sorts the variables in order of decreasing bounds. Given a node (S, k), the bound of the
ith active variable (i = 1, . . . , d) is RSS(drop(S, k + i)), i.e. the RSS of the model from which the (k + i)th variable
has been removed. The d active variables of the node (S, k) are represented by the leading d × d submatrix of the
upper triangular R ∈ R(d+1)×(d+1) factor of the QR decomposition. The last column of R corresponds to the response
22
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
Fig. 4. Exploiting the QR decomposition to compute the bound of the ith active variable, when i = 2.
variable y in (1). Let R̃ denote R without its ith column. The drop operation applies d − i + 1 biplanar Givens rotations
to retriangularize R̃. That is, it computes
R̂ d
Gd · · · Gi R̃ =
,
0 1
(5)
where R̂ ∈ Rd×d is upper triangular. The bound of the ith active variable, that is, the RSS of the model after deleting
2 —the square of the diagonal element of R̂ in position (d, d).
the ith variable, is given by R̂d,d
Note that the rotation Gj R̃ (j = i, . . . , d) affects only the last (d − j + 1) elements of rows j and j + 1 of
R̃, and reduces to zero R̃j +1,j . The application of a rotation to two biplanar elements x and y can be written
as
x̃
c
=
ỹ
−s
s
c
x
.
y
If c and s are chosen such that c = x/t and s = y/t, then x̃ = t and ỹ = 0, where t 2 = x 2 + y 2 and t = 0.
The number of nodes in which the variables are preordered increases exponentially with the preordering radius
P. This computational overhead will have a significant impact on the overall performance of the RadiusBBA.
Fig. 4(a) shows the retriangularization of a 6 × 6 triangular matrix after deleting the second column using Givens
rotations.
The complete and explicit triangularization of R̃ should be avoided and only the bound of the deleted ith variable,
2 , should be computed. This can be achieved by computing only the elements of R̃ which are needed in deriving
i.e. R̂d,d
R̂d,d . Thus, the Givens rotation Gj R̃ (j = i, . . . , d + 1) explicitly updates only the last (d − j + 1) elements of the
(j + 1)th row of R̃ which are required by the subsequent rotation. The jth row of R̃ is not updated and neither is the
subdiagonal element R̃j +1,j annihilated. This strategy has approximately half the cost for deriving the bound of the
ith variable. In order to optimize the implementation of this procedure, the original triangular R is not modified and
the bounds are computed without copying the matrix to temporary store. Fig. 4(b) illustrates the steps of this strategy,
while Algorithm 3 shows the exact computational steps.
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
30
400
25
execution time (s)
350
generated nodes
23
300
250
200
20
15
10
5
150
0
2
4
6
8
10
preordering radius
12
14
0
(a) POLLUTE dataset (number of nodes)
10
20
30
preordering radius
40
50
(b) 13 variable true model (execution time)
1200
100
execution time (s)
execution time (s)
1000
80
60
40
800
600
400
20
200
0
10
20
30
preordering radius
40
50
(c) 26 variable true model (execution time)
0
10
20
30
preordering radius
40
50
(d) 39 variable true model (execution time)
Fig. 5. Computational cost of RadiusBBA for the POLLUTE data set and artificial data with 52 variables.
Algorithm 3. Computing the bound of the ith active variable
1
2
3
4
5
6
7
8
9
procedure bound(R, i, b)
xj ← Ri+1,j , for j = i + 1, . . . , d + 1
for j = i + 1, . . . , d + 1 do
yk ←Rj +1,k , k = j, . . . , d + 1
t ← xj2 + yj2 ; c ← xj /t; s ← yj /t
xk ← −s · xk + c · yk , k = j + 1, . . . , d + 1
end for
b ← t2
end procedure
Fig. 5 illustrates the effect of the preordering radius P on the RadiusBBA. Fig. 5(a) illustrates the number of nodes
generated by the RadiusBBA on the POLLUTE data set (15 variables) for every preordering radius (1 P 15). The
BBA-1 generates 318 nodes. The number of nodes generated by the RadiusBBA decreases steadily up to P = 8 where a
minimum of 146 nodes is reached. The other three figures illustrate the execution times of the RadiusBBA on artificial
data sets with 52 variables. The true model comprises 13, 26 and 39 variables, respectively. In all three cases the
RadiusBBA represents a significant improvement over the BBA-1. In the case of the small true model (13 variables),
24
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
Table 3
Execution times in seconds of the subrange RadiusBBA with radius 21 and 26 on data sets comprising 64 and 80 variables, respectively
ntrue
n = 64
16
32
48
n = 80
20
40
60
na
nb
Time
1
8
1
24
1
36
64
24
64
40
64
60
119
50
3415
4
3531
1
1
10
1
20
1
40
80
30
80
60
80
80
4205 (70 min)
2309 (38 min)
177383 (2 days)
25732 (8 h)
1293648 (15 days)
178 (3 min)
The true model comprises ntrue variables.
the BBA-1 and the RadiusBBA with P = 9 require 30 and 1 s, respectively. In the case of a medium size true model
(26 variables), the time is reduced from 112 to 6 s, i.e. the RadiusBBA with radius 13 is almost 20 times faster. In
the third case (big true model with 39 variables) the RadiusBBA with radius 18 is over 2 times faster than the BBA-1
which requires 500 s. These tests show empirically that values of P lying between n/4 and n/3 are a good choice for
the RadiusBBA.
Table 3 shows the execution times of the RadiusBBA on two data sets with n = 64 and 80 variables, respectively.
The preordering radius used is P = n/3. The number of variables in the true model is given by ntrue . Different ranges
na and nb have been used. For the full range, i.e. na = 1 and nb = n, the RadiusBBA computes the best subset models
for model sizes which are computationally infeasible for the BBA-1. It can be observed that the use of smaller ranges
significantly reduces the time required by the RadiusBBA for deriving the best subset models.
4. Heuristic strategies
The Heuristic BBA (HBBA) relaxes the objective of finding an optimal solution in order to gain in computational efficiency. That is, the HBBA is able to tackle large-scale models when the exhaustive BBA is found to be computationally
infeasible. The heuristic algorithm ensures that
RRE(S̃i ) < for i = 1, . . . , n,
(6)
where Si is the (heuristic) solution subset model of size i and is a tolerance parameter ( > 0). Generally, the RRE of
a subset Si is given by
RRE(Si ) =
|RSS(Si ) − RSS(S∗i )|
,
RSS(S∗i )
where Si∗ is the optimal subset of size i reported by the BBA. The space of all possible submodels is not searched
exhaustively. The HBBA aims to find an acceptable compromise between the brevity of the search ( → ∞) and the
quality of the solutions computed ( → 0). The modified cutting test in Gatu and Kontoghiorghes (2006) is given by
(1 + ) · RSS(S) > rj +1 .
(7)
Note that the HBBA is equivalent to the BBA for = 0. Furthermore, the HBBA reduces to the DCA if = −1.
Notice that in (7) rj +1 < 0 for = −1, which implies that the cutting test never holds and all the nodes of the tree are
generated.
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
25
λ(i)
2τ
τ
0
0
n−1
1 2
level i
Fig. 6. The (i) level tolerance function.
Table 4
Mean number of nodes and RREs generated by the HBBA and LevelHBBA
ntrue
9
18
27
Algorithm
Nodes
RRE
Nodes
RRE
Nodes
RRE
HBBA
LevelHBBA
14 278
13 129
6e − 4
8e − 4
47 688
34 427
3e − 4
5e − 4
35 062
21 455
9e − 4
3e − 3
In order to increase the capability of the heuristic strategy to tackle larger subset-selection problems, a new heuristic
algorithm is proposed. The Level HBBA (LevelHBBA) employs different values of the tolerance parameter in different
levels of the regression tree. It uses higher values in the levels close to the root node to encourage the cutting of large
subtrees. Lower tolerance values are employed in lower levels of the tree in order to select good quality subset models.
The indices of the tree levels are shown in Fig. 1. The tolerance function employed by the LevelHBBA is defined
formally as
(i) = 2(n − i − 1)/(n − 1)
for i = 0, . . . , n − 1,
where i denotes the level of the regression tree and the average tolerance. The graph of the function (i) is shown in
Fig. 6.
The HBBA and LevelHBBA were executed on data sets with 36 variables. Three types of data sets were employed,
with a small, a medium and a big true model comprising 9, 18 and 27 variables, respectively. The tolerance parameter has been set to 0.2. The results are summarized in Table 4. The table shows the number of nodes and the mean RRE. Each
experiment has been repeated 32 times. The values shown in the table are overall means. The LevelHBBA generates
slightly fewer nodes, but it produces results that are of lesser quality than those computed by the HBBA. Notice that
the average RRE is significantly lower than the tolerance employed.
The Size HBBA (SizeHBBA) assigns a different tolerance value to each subset model size. It can be seen as a
generalization of the HBBA and the RangeBBA. The degree of importance of each subset size can be expressed.
Lower tolerance values are attributed to subset sizes of greater importance. Less relevant subset sizes are given higher
tolerance values. Subset model sizes can be effectively excluded from the search by setting a very high tolerance
value. Thus, unlike the RangeBBA, the SizeHBBA can be employed to investigate non-contiguous size ranges. The
SizeHBBA satisfies
RRE(S̃i ) i
for i = 1, . . . n,
where i denotes the size of the subset model and i the corresponding tolerance value. Given a node (S, k), the child
node (drop(S, j ), j − 1) is cut if:
(1 + i ) · RSS(S) > ri
for i = j, . . . , ns − 1.
26
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
Table 5
Mean number of nodes and RREs generated by the HBBA and SizeHBBA
ntrue
9
18
27
Algorithm
Nodes
RRE
Nodes
RRE
Nodes
RRE
HBBA
SizeHBBA
12 781
15 079
8e − 4
2e − 4
38 716
39 457
4e − 4
2e − 4
39 907
40 250
1e − 3
3e − 4
The SizeHBBA generalizes the previous algorithms, i.e.,
⎧
DCA
if i = −1,
⎪
⎪
⎪ RangeDCA if i = −1 for na i nb and i ?0 otherwise,
⎨
SizeHBBA ≡ BBA
if i = 0,
⎪
⎪
⎪
⎩ RangeBBA if i = 0 for na i nb and i ?0 otherwise,
HBBA
if i = .
The SizeHBBA is equivalent to all previously proposed algorithms with the exception of the LevelHBBA. Thus, it can
be seen as more than a mere heuristic algorithm and allows a very flexible investigation of all subset models.
It has been observed experimentally that the SizeHBBA is efficient compared to the HBBA when a tolerance value
is used for the first half of model sizes and zero-tolerance for the remaining sizes. That is, when the optimal solution
is guaranteed to be found for submodel sizes between n/2 and n. Table 5 shows the computational performance of the
HBBA and SizeHBBA on data sets with 36 variables. The HBBA is executed with = 0.2, while the SizeHBBA is
executed with i = for i 18 and i =0 otherwise. The results show that, without a significant increase in computational
cost, there is a gain in solution quality, i.e. the optimal subset models with 18 or more variables are found. Furthermore,
the results are consistent with the observed behavior of the RangeBBA. For bigger models, larger subranges can be
chosen at a reasonable computational cost. In case of the SizeHBBA, constraints on larger submodels can be stricter
(i.e. lower tolerance) without additional computational cost. This may be due to the asymmetric structure of the tree,
i.e. subtrees are smaller on the right hand side.
5. Conclusions
Various algorithms for computing the best subset regression models have been developed. They improve and extend
previously introduced exhaustive and heuristic strategies which were aimed at solving large-scale model-selection
problems. The proposed algorithms are based on a dropping column algorithm (DCA) which derives all possible subset
models by generating a regression tree (Gatu and Kontoghiorghes, 2003, 2006; Smith and Bremner, 1989).
An algorithm (RangeDCA) that computes the all-subsets models within a given range of model sizes has been
proposed. The RangeDCA is a generalization of the DCA and it generates only a subtree of the all-subsets tree derived
by the DCA. Theoretical measures of complexity of the RangeDCA have been derived and analyzed (see Appendix).
The theoretical complexities have been confirmed through experiments. The branch-and-bound strategy in Gatu and
Kontoghiorghes (2006) has been applied in the tree that is generated by the RangeDCA.
The preordering of the initial variable set (BBA-1) significantly improves the computational performance of the
BBA. However, the BBA-1 might fail to detect significant combinations of variables in the root node. Hence, a more
robust preordering strategy is designed. Subsets of variables are sorted inside the regression tree after some variables
have been deleted. Thus, important combinations of variables are more likely to be identified and exploited by the
algorithm.
A preordering BBA (RadiusBBA) which generalizes the BBA-1 has been designed. The RadiusBBA applies variable preordering to nodes of arbitrary radius in the regression tree rather than to the root only. The radius provides a
measure of the distance between a node and the root. Experiments have shown that the number of nodes computed
by the RadiusBBA decreases as the preordering radius increases. However, the preordering requires the retriangularization of an upper triangular matrix after deleting a column and it incurs a considerable computational overhead. A
computationally efficient strategy has been designed, which avoids the explicit retriangularization used to compute the
strength of a variable. This reduces the total overhead of the RadiusBBA. In various experiments, it has been observed
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
27
that the best performance is achieved when preordering is employed with a radius of between one quarter and one third
of the number of variables. The RadiusBBA significantly reduces the computational time required to derive the best
submodels when compared to the existing BBA-1. This allows the RadiusBBA to tackle subset-selection problems that
have previously been considered as computationally infeasible.
A second class of algorithms has been designed, which improve the heuristic version of the BBA (HBBA) (Gatu
and Kontoghiorghes, 2006). The Level HBBA (LevelHBBA) applies different tolerances on different levels of the
regression tree. The LevelHBBA generates fewer nodes than the HBBA when both algorithms are applied with the
same mean tolerance. Although the subset models computed by the LevelHBBA are of lesser quality than those
computed by the HBBA, the relative residual errors remain far below the mean tolerance. The size-heuristic BBA
(SizeHBBA) assigns different tolerance values to subset models of different sizes. The subset models computed by the
SizeHBBA improve the quality of the models derived by the HBBA. Thus, for approximately the same computational
effort, the SizeHBBA produces submodels closer to the optimal ones than does the HBBA. The SizeHBBA for different
kind of tolerances is equivalent to the DCA, RangeDCA, BBA, RangeBBA and HBBA. This makes the SizeHBBA a
powerful and flexible tool for computing subset models. Within this context, it extends the RangeBBA by allowing the
investigation of submodels of non-contiguous size ranges.
The employment by the RadiusBBA of computationally less expensive criteria in preordering the variables should be
investigated. This should include the use of parallel strategies to compute the bound of the model after deleting a variable
(Hofmann and Kontoghiorghes, 2006). It might be fruitful to explore the possibility of designing a dynamic heuristic
BBA which automatically determines the tolerance value in a given node based on a learning strategy. A parallelization
of the BBA, employing a task-farming strategy on heterogeneous parallel systems, could be considered. The adaptation
of the strategies to the vector autoregressive model is currently under investigation (Gatu and Kontoghiorghes, 2005,
2006).
Acknowledgments
The authors are grateful to the guest-editor Manfred Gilli and the two anonymous referees for their valuable comments
and suggestions. This work is in part supported by the Swiss National Science Foundation Grants 101412-105978,
200020-100116/1, PIOI1-110144 and PIOI1-115431/1, and the Cyprus Research Promotion Foundation Grant KYIT/0906/09.
Appendix A. Subrange model selection: complexity analysis
Let the pair (S, k) denote a node of the regression tree, where S is a set of n variables and k the number of passive
variables (0 k < n). A formal representation of the DCA regression tree is given by
(S, k)
if k = n − 1,
(S, k) =
((S, k), (drop(S, k + 1), k), . . . , (drop(S, n − 1), n − 2)) if k < n − 1.
The operation drop(S, i) deletes the ith variable in S = [s1 , . . . , sn ]. The QR decomposition is downdated after the
corresponding column of the data matrix has been deleted. Orthogonal Givens rotations are employed in reconstructing
the upper-triangular factor. An elementary operation is defined as the rotation of two vector elements. The cost of
one elementary operation is approximately six flops. The number of elementary operations required by the drop
operation is
Tdrop (S, i) = (n − i + 1)(n − i + 2)/2.
The passive variables s1 , . . . , sk are not dropped, i.e. they are inherited by all child nodes. All active variables
sk+1 , . . . , sn , except the last one, are dropped in turn to generate new nodes. The structure of the tree can be expressed in terms of the number of active variables d = n − k. This simplified representation (d) of the regression tree
(S, k) is given by
(d)
if d = 1,
(d) =
((d), (d − 1), . . . , (1)) if d > 1,
28
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
where (d) is a node with d active variables. The number of nodes and elementary operations are calculated,
respectively, by
N(d) = 1 +
d−1
N(d − i) = 2d−1
i=1
and
T (d) =
d−1
(Tdrop (d, i) + T (d − i)) = 7 · 2d−1 − (d 2 + 5d + 8)/2.
i=1
Here, Tdrop (d, i) is the complexity of dropping the ith of d active variables (i = 1, . . . , d).
Let na designate a model size (1na n). Then, na (S, k) denotes the subtree of (S, k) which consists of all nodes
which evaluate exactly one model of size na (0 k < na ). It is equivalent to:
(d)
if d = a,
a (d) =
((d), a (d − 1), . . . , 1 (d − a)) if d > a,
where a = na − k. The number of nodes is calculated by
1 if d = a,
Na (d) =
1 + ai=1 Na−i+1 (d − i) if d > a
d!
= Cad =
.
a!(d − a)!
Similarly, the number of elementary operations required to construct a (d) is calculated by
0
if d = a,
Ta (d) = a
i=1 (Tdrop (d, i) + Ta−i+1 (d − i)) if d > a
= Tdrop (d, 1) + Ta−1 (d − 1) + Ta (d − 1).
The closed form
Ta (d) =
a−1
d−a+i−1
i=0
j =i
j
Ci · Tdrop (d − j, 1)
is obtained through the generating function
Tdrop (j, i)x i y j
G(x, y) = (1 − y(1 + x))−1
0<i<j
of Ta (d), where k = 0, a = na and d = n. That is, this is the number of elementary operations necessary to compute
all subset models comprising na out of n variables.
Now, let na ,nb (S, k) denote the tree which evaluates all subset models with more than na and less than nb variables,
inclusively (1 na nb n and 0 k < na ). It is equivalent to
(d)
if d = a,
a,b (d) = a,b−1 (d)
if d = b,
((d), a,b (d − 1), . . . , 1,b−a+1 (d − a), . . . , 1,1 (d − b)) if d > b,
where a = na − k and b = nb − k. This tree can be seen as the union of all trees c (d), for c = a, . . . , b. Hence, the
number of nodes and operations can be calculated, respectively, by
Na,b (d) =
b
c=a
Nc (d) −
b−1
c=a
Nc (d)
M. Hofmann et al. / Computational Statistics & Data Analysis 52 (2007) 16 – 29
29
and
Ta,b (d) =
b
Tc (d) −
b−1
Tc (d).
c=a
c=a
Now,
Nc (d) = Ccd−1
and
Tc (d) =
c−1
d−c+i−2
i=0
j =i
j
Ci · Tdrop (d − j, 1)
are the nodes and operations which have been counted twice. Specifically, these are given by the subtrees nc (S, k)
which represent the intersection of the two trees nc (S, k) and nc +1 (S, k) for 1 nc < n. Their structure is given by
(d)
if d = c + 1,
c (d) =
((d), c (d − 1), . . . , 1 (d − a)) if d < c + 1,
where c = nc − k.
References
Breiman, L., 1995. Better subset regression using the nonnegative garrote. Technometrics 37, 373–384.
Clarke, M.R.B., 1981. Statistical algorithms: algorithm AS 163: a Givens algorithm for moving from one linear model to another without going back
to the data. J. Roy. Statist. Soc. Ser. C Appl. Statist. 30, 198–203.
Fan, J., Li, R., 2001. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Amer. Statist. Assoc. 96, 1348–1360.
Furnival, G., Wilson, R., 1974. Regression by leaps and bounds. Technometrics 16, 499–511.
Gatu, C., Kontoghiorghes, E.J., 2003. Parallel algorithms for computing all possible subset regression models using the QR decomposition. Parallel
Comput. 29, 505–521.
Gatu, C., Kontoghiorghes, E.J., 2005. Efficient strategies for deriving the subset VAR models. Comput. Manage. Sci. 2, 253–278.
Gatu, C., Kontoghiorghes, E.J., 2006. Branch-and-bound algorithms for computing the best subset regression models. J. Comput. Graph. Statist. 15,
139–156.
Gatu, C., Yanev, P., Kontoghiorghes, E.J., 2007. A graph approach to generate all possible regression submodels. Comput. Statist. Data Anal. in
press, doi: 10.1016/j.csda.2007.02.018.
Golub, G.H., Van Loan, C.F., 1996. Matrix Computations. Johns Hopkins Studies in the Mathematical Sciences. third ed. Johns Hopkins University
Press, Baltimore, MA.
Hastie, T., Tibshirani, R., Friedman, J., 2001. The Elements of Statistical Learning. Springer Series in Statistics. Springer, New York.
Hocking, R.R., 1976. The analysis and selection of variables in linear regression. Biometrics 32, 1–49.
Hofmann, M., Kontoghiorghes, E.J., 2006. Pipeline givens sequences for computing the QR decomposition on a EREW PRAM. Parallel
Comput. 32, 222–230.
Kontoghiorghes, E.J., 2000. Parallel Algorithms for Linear Models: Numerical Methods and Estimation Problems, Advances in Computational
Economics, vol. 15. Kluwer Academic Publishers, Boston.
Miller, A.J., 2002. Subset Selection in Regression Monographs on Statistics and Applied Probability, vol. 95, second ed. Chapman & Hall, London
(Related software can be found at URL: http://users.bigpond.net.au/amiller/).
Narendra, P.M., Fukunaga, K., 1997. A branch and bound algorithm for feature subset selection. IEEE Trans. Comput. 26, 917–922.
R Development Core Team, 2005. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna,
Austria.
Roberts, S.J., 1984. Statistical algorithms: algorithm AS 199: a branch and bound algorithm for determining the optimal feature subset of given size.
Appl. Statist. 33, 236–241.
Searle, S.R., 1971. Linear Models. Wiley, New York.
Seber, G.A.F., 1977. Linear Regression Analysis. Wiley, New York.
Sen, A., Srivastava, M., 1990. Regression Analysis. Theory, Methods and Applications. Springer, Berlin.
Smith, D.M., Bremner, J.M., 1989. All possible subset regressions using the QR decomposition. Comput. Statist. Data Anal. 7, 217–235.
Somol, P., Pudil, P., Kittler, J., 2004. Fast branch & bound algorithms for optimal feature selection. IEEE Trans. Pattern Anal. Mach. Intell. 26,
900–912.
Tibshirani, R.J., 1996. Regression shrinkage and selection via the lasso. J. Roy. Statist. Soc. Ser. B (Statist. Methodol.) 58, 267–288.
An exact least-trimmed-squares algorithm for a range of
coverage values
Marc Hofmann∗,
Cristian Gatu†,
Erricos John Kontoghiorghes‡
Abstract
A new adding row algorithm (ARA) extending existing methods to compute exact least trimmed squares
(LTS) regression is presented. The ARA employs a tree-based strategy to compute LTS regressors for a range
of coverage values. Thus, a priori knowledge of the optimal coverage parameter is not required. New nodes
in the regression tree are generated by updating the QR decomposition after adding one observation to the
regression model. The ARA is enhanced employing a branch-and-bound strategy. The branch-and-bound
algorithm is an exhaustive algorithm that uses a cutting test to prune non-optimal subtrees. It significantly
improves over the ARA in computational performance. Observation preordering throughout the traversal
of the regression tree is investigated. A computationally efficient and numerically stable calculation of the
bounds using Givens rotations is designed around the QR decomposition, avoiding the need to explicitely
update the triangular factor when an observation is added. This reduces the overall computational load
of the preordering device by approximately half. A solution is proposed to allow preordering during the
execution of the algorithm when the model is underdetermined. It employs pseudo-orthogonal rotations to
downdate the QR decomposition. The strategies are illustrated by example. Experimental results confirm
the computational efficiency of the proposed algorithms.
Keywords: Least trimmed squares, outliers, regression tree algorithms, QR factorization.
1
Introduction
Least-squares regression is sensitive to outliers. This has prompted the search for regression estimators which
are resistant to data points that deviate from the usual assumptions. The goal of positive-breakdown estima∗
†
Institut d’informatique,Université de Neuchâtel, Switzerland. E-mail: marc.hofmann@unine.ch
VTT Technical Research Centre of Finland, Espoo, Finland; and Faculty of Computer Science, “Alexandru Ioan Cuza” Univer-
sity of Iaşi, Romania. E-mail: cristian.gatu@unine.ch
‡
Department of Public and Business Administration, University of Cyprus, Cyprus; and School of Computer Science and Information Systems, Birkbeck College, University of London, UK. E-mail: erricos@ucy.ac.cy
1
tors is to be robust against the possibility of unannounced outliers (Rousseeuw 1997). The breakdown point
provides a crude quantification of the robustness properties of an estimator. Briefly, the breakdown point is
the smallest amount of contamination that may cause an estimator to take on arbitrarily large aberrant values
(Donoho & Huber 1983).
Consider the standard regression model
y = Xβ + ε,
(1)
where y ∈ Rn is the dependent-variable vector, X ∈ Rn×p is the exogenous data-matrix of full column-rank,
β ∈ Rp is the coefficient vector and ε ∈ Rn is the noise vector. It is usually assumed that ε is normally
distributed with zero mean and variance-covariance matrix σ 2 In . Least squares (LS) regression consists of
minimizing the residual sum of squares (RSS). One outlier may be sufficient to compromise the LS estimator.
In other words, the finite-sample breakdown point of the LS estimator is 1/n and therefore tends to 0 when n is
large (Rousseeuw 1997). Several positive-breakdown methods for robust regression have been proposed, such
as the least median of squares (LMS) (Rousseeuw 1984). The LMS is defined by minimizing medi ε̂2i . The
LMS attains the highest-possible breakdown-value, namely (⌊(n − p)/2⌋ + 1)/n. This means that the LMS
fit stays in a bounded region whenever ⌊(n − p)/2⌋, or fewer, observations are replaced by arbitrary points
(Rousseeuw & Van Driessen 2006).
The least trimmed squares (LTS) estimator possesses better theoretical properties than the LMS (Rousseeuw
& Van Driessen 2006, Hössjer 1994). The objective of the LTS estimator is to minimize
h
X
hε̂2 ii ,
i=1
where h = 1, . . . n and hε̂2 i denotes the vector of squared residuals sorted in increasing order. This is equivalent
to finding the h-subset of observations with the smallest LS objective function. The LTS regression estimate
is then the LS fit to these h points. The breakdown value of LTS with h = ⌊(n + p + 1)/2⌋ is equivalent to
that of the LMS. In spite of its advantages over the LMS estimator, the LTS estimator has been applied less
often because it is computationally demanding. For the multiple linear regression model, the trivial algorithm
that explicitly enumerates and computes the RSS for all h-subsets works if the number of observations is
relatively small, i.e. less than 30. Otherwise, the computational load is prohibitive. To overcome this drawback,
several approximate algorithms have been proposed. These include the PROGRESS algorithm (Rousseeuw &
Leroy 1987), the feasible-solution-algorithm (FSA) (Hawkins 1994, Hawkins & Olive 1999) and the FASTLTS algorithm (Rousseeuw & Van Driessen 2006). However, these algorithms do not inspect all combinations
of observations and are not guaranteed to find the optimal solution. An exact algorithm to calculate the LTS
estimator has been proposed (Agulló 2001). It is based on a branch-and-bound procedure that does not require
the explicit enumeration of all h-subsets. It therefore reduces the computational load of the trivial algorithm
2
significantly. A tree-based algorithm to enumerate subsets of observations in the context of outlier detection
has been suggested (Belsley, Kuh & Welsch 1980).
High-breakdown estimators may pick up local linear trends with different slopes than the global linear trend.
Thus, high-breakdown estimators may have arbitrarily low efficiency (Stefanski 1991, Morgenthaler 1991).
For higher values of the breakdown point the probability for this to happen is bigger. Thus, for small n/p it
is preferable to use a method with lower breakdown value such as the LTS with larger h (Rousseeuw 1997).
Here, an adding row algorithm (ARA) is proposed which computes the exact LTS estimates for a range hmin :
hmax = {h ∈ N|h ≥ hmin , h ≤ hmax }. It renders possible the efficient computation and investigation of a set of
exact LTS estimators for distinct breakdown values h = hmin , . . . , hmax . A branch-and-bound algorithm (BBA)
improves upon the ARA. Its computational efficiency is further improved by preordering the observations.
The ARA and its application to LTS regression is discussed in Section 2. The BBA is introduced in Section 3, observation preordering is investigated and numerical results are presented. Conclusions and notions of
future work are discussed in Section 4 .
2
Adding row algorithm
As noted by Belsley et al. (1980), there is a strong correspondence between row-selection techniques and
procedures for computing the all-possible-column-subsets regression. Within the context of variable-subset
selection, a Dropping column algorithm (DCA) has been discussed (Gatu & Kontoghiorghes 2003, Gatu &
Kontoghiorghes 2006, Smith & Bremner 1989). The new Adding row algorithm (ARA) computes the allobservation-subsets regression. The organization of the algorithm is similar to that of the DCA and is determined by the all-subsets tree illustrated in Figure 1 (Furnival & Wilson 1974, Gatu & Kontoghiorghes 2003,
Smith & Bremner 1989), where the number of observations in the model is n = 4.
[], [1234]
level 0
[1], [234]
1
2
[12], [34]
3
[123], [4]
4
[1234], []
[124], []
[13], [4]
[2], [34]
[14], []
[134], []
[23], [4]
[24], []
[234], []
Figure 1: The ARA regression tree where I = [1234].
3
[3], [4]
[34], []
[4], []
The observations or points in model (1) are designated by their indices I = [1, . . . , n]. A node (S, A) in the
regression tree carries an observation-subset model of nS selected observations S ⊆ I. The set A represents
the nA observations which are available for selection in nodes that are children of node (S, A). The RSS of the
LS estimator that corresponds to the subset model S is computed in each node. The subset model is denoted by
(XS yS ) and is assumed to be of full rank. The regression model is represented by means of the numerically
stable QR decomposition (QRD)
QTS
p
1
XS yS
=
p
1
RS zS
0
w
p
,
(2)
nS −p
where QS is orthogonal and RS is square upper-triangular and non-singular. The RSS of the model is given by
ρS = wT w. Note that for underdetermined models (nS < p), the QR factorization is not computed and the RSS
is 0. The orthogonal factor QTS is typically a product of Givens rotations, or Householder reflectors (Golub &
Van Loan 1996).
Given any node (S, A = [a1 , . . . , anA ]), its nA child nodes are given by
[add(S, a1 ), A2: ),
(add(S, a2 ), A3: ),
...,
(add(S, anA ), ∅)] ,
where Ai: denotes the subset of A containing all but the i − 1 first observations. The operation add(S, ai ),
i = 1, . . . , nA , constructs the new linear model S ∪ {ai } by updating the linear model S with observation
ai . Effective algorithms to update the quantities RS , zS and ρS after adding an observation exist (Gill, Golub,
Murray & Saunders 1974). A Cholesky updating routine based on orthogonal Givens rotations can be found
in the LINPACK numerical library (Dongarra, Bunch, Moler & Stewart 1979). It requires approximately 3p2
flops (Björck, Park & Eldén 1994).
A straight forward “brute-force” (BF) method to compute the exact LTS estimator β̂h for a given coverage
h (h = 1, . . . , n) consists in enumerating all possible h-subsets, solving the LS problem for each subset. This
implies computing nh = n!/(h!(n − h)!) QRDs. Thus, the computational cost amounts to approximately
TBF = nh · 3hp2 flops, where 3hp2 is the approximate number of flops necessary to compute the QRD. On the
other hand, the specialized algorithm ARAh to compute the LTS regressor β̂h for a given coverage h lists the
observation subsets in an order predetermined by the all-subsets tree. Starting at the root node, it traverses all
nodes that lead to a node on level h. The nodes on level h are included in the traversal. Although it enumerates
more subsets than the BF algorithm, the computational load is less. By exploiting the information gathered in
intermediate nodes, the QRDs are obtained cheaply. That is, the QRD in a node on level ℓ is partially available
as the QRD in the parent node (level ℓ − 1). The new QRD is not derived from scratch. Instead, it is obtained
by adding an observation to the selected set S. Numerically, this implies updating the parent QRD by one row.
4
Let ∆(S, A) denote the all-observation-subsets tree with root node (S, A) and let ∆h (S, A) denote the tree
generated by the ARAh . The tree ∆h (S, A) is the subtree of ∆(S, A) that contains the nodes that lie on a path
between the root node and a node on level h. It is equivalent to the tree employed in feature subset regression
by Narendra & Fukunaga (1977) and by Agulló’s (2001) exact LTS algorithm (hereafter denoted by AGLA).
Formally,
∆h (S, A) =
∅
if
nS + nA < h or
nS > h,
[(S, A), ∆h (add(S, a1 ), A2: ), . . . , ∆h (add(S, an ), ∅)]
A
otherwise,
nodes, where n = |I|. Thus, the
where nS = |S| and nA = |A|. The tree ∆h (∅, I) consists of n+1
h
n+1
computational cost of the ARA is approximately TARAh = h · 3p2 flops. In other words,
TBF /TARAh ≈ α(1 − α)n + α = O(n),
where n ≫ p and h = αn (α < 1). The ARAh is O(n) times faster than the brute-force method.
The optimal value of h will both resist outliers in the data and give the highest efficiency, i.e. will accurately reveal the global linear trend. However, in practice, this value is never known before using LTS regression
(Atkinson & Cheng 1999). The ARAh visits all the nodes in ∆h to determine β̂h , the LTS estimator with coverage h. Several passes are required to find all the LTS estimators β̂h , h = hmin , . . . , hmax . Each pass executes
P max
n+1
nodes. Notice that nodes will be computed several
the ARAh for one h. This implies computing hh=h
h
min
times over. It is inefficient to analyse the same data several times for different vaues of h.
Now, let ∆hmin :hmax (S, A) denote the subtree of ∆(S, A) that contains all nodes that lie on a path between
the root node and a node on level ℓ (ℓ = hmin , . . . , hmax ). Formally,
∅ if nS + nA < hmin or nS > hmax ,
∆hmin :hmax (S, A) =
[(S, A), ∆h :h (add(S, a1 ), A2: ), . . . , ∆h :h (add(S, an ), ∅)]
min max
min max
A
otherwise.
The ARAhmin :hmax generates the tree ∆hmin :hmax (∅, I) and returns the set of LTS estimators {β̂h |h = hmin , . . . , hmax }.
It is optimal in the sense that it does not generate any unnecessary nodes and that it does not generate any node
multiple times. The number of nodes that it computes is given by
NARA =
hX
hmax max −1 X
n
n+1
,
−
h
h
h=h
h=h
min
min
thus improving over the previous approach. The complete procedure for generating ∆hmin :hmax (∅, I) is given by
Algorithm 1. Nodes which await processing are held in a node list. The list is managed according to a LIFO
(“last in, first out”) strategy (Burks, Warren & Wright 1954, Newell & Shaw 1957). The output of the algorithm
is ρ̂h and Ŝh , respectively the RSS of the LTS estimate β̂h and the corresponding h-subset (h = hmin , . . . , hmax ).
5
Algorithm 1: Adding row algorithm (ARA)
1
procedure ARA(I, hmin , hmax , ρ̂, Ŝ)
2
n ← |I|
3
ρ̂h ← +∞, where h = hmin , . . . , hmax
4
Insert node (∅, I) into node list
5
while list contains nodes do
6
Remove node (S, A) from node list
7
nS ← |S|; nA ← |A|; ρS ← RSS(S)
8
if nS ≥ hmin and nS ≤ hmax then
9
if ρS < ρ̂nS then ρ̂nS = ρS , ŜnS ← S
10
end if
11
if nS + nA ≥ hmin and nS < hmax then
12
for i = 1, . . . , nA do
13
Compute (S ′ , A′ ) = (add(S, ai ), Ai+1: )
14
Insert (S ′ , A′ ) into node list
15
end for
16
end if
17
end while
18
end procedure ARA
Hofmann, Gatu & Kontoghiorghes (2007) give a detailed analysis of the tree structures and the associated
complexities in the context of variable-subset selection.
The advantage of the ARA over the previously introduced AGLA is that the ARA simulataneously computes the LTS estimators for a range of coverage values 1 ≤ hmin , . . . , hmax ≤ n. It traverses the tree ∆hmin :hmax
only once and avoids redundant computations. In contrast, the AGLA can determine only one LTS regressor at
a time. Thus, in order to obtain the same list of LTS estimators, the AGLA is executed hmax − hmin + 1 times.
This implies several independent traversals of the subsets-tree and the computation of redundant nodes.
2.1 Experimental results
To see the effect of the coverage parameter h on the LTS estimator, the ratio suggested by Atkinson & Cheng
(1999) is considered:
Rh =
6
σ̂h2
,
2
σ̂LS
where σ̂h2 = ρ̂h /(h − p) and ρ̂h = RSS(β̂h ). The variance estimate associated with the LS estimator is given
2
by σ̂LS
= σ̂n2 . The ratio Rh is examined as different numbers of data are fitted in the model. Two data models
are used in the experiment. Atkinson & Cheng (1999) used the model
yi = β0 +
p−1
X
βj xi,j + εi ,
i = 1, . . . , n,
j=1
where εi ∼ N (0, 1) for good data, whereas bad data are simulated from N (12, 1). Rousseeuw & Van Driessen
(2006) used the model
yi = xi,1 + xi,2 + . . . + xi,p−1 + 1 + εi ,
i = 1, . . . , n,
where εi ∼ N (0, 1) and xi,j ∼ N (0, 100). Outliers are introduced by replacing some of the xi,1 by values that
are normally distributed with mean 100 and variance 100.
Two classes of datasets are generated, one for each data model. Each class contains 100 datasets with
n = 32 and p = 5. The contaminated datasets are obtained from a clean dataset by injecting q = 8 outliers.
The ARA is then executed on all 100 datasets with hmin = 16 and hmax = 32, and the results reported for
both clases of data. For the first class of data (Atkinson & Cheng 1999), the ARA correctly discriminated the
contaminated observations in 92 of 100 cases. This means that for 92 out of 100 contaminated datasets, the
estimator β̂n−q=24 did not include any of the contaminated data points. For the second class of data (Rousseeuw
& Van Driessen 2006), the ARA correctly discriminated the outliers in all 100 cases. Figure 2 illustrates the
ratio Rh for the two classes of data. Each plot shows the ratio Rh for one case, i.e. a dataset with and without
1.4
1.0
0.8
R(h)
0.0
0.2
0.4
0.6
1.0
0.8
0.6
0.4
0.2
0.0
R(h)
clean
contaminated
1.2
clean
contaminated
1.2
1.4
contamination.
20
25
30
20
coverage (h)
25
30
coverage (h)
(a) Data: Atkinson & Cheng (1999).
(b) Data: Rousseeuw & Van Driessen (2006).
Figure 2: The ratio Rh for two types of data.
7
3
Branch and bound algorithm
The ARA is computationally prohibitive for even a moderate number of observations to investigate. The ARA
can be optimized to avoid the explicit enumeration of all observation-subsets. Given two sets of observations
S1 and S2 ,
if
S1 ⊂ S2 , then
ρS 1 ≤ ρS 2 ,
where ρS denotes the RSS of the LS estimator of the model consisting of the observations in S. That is, adding
observations cannot cause the RSS of the model to decrease. This property can be used to restrict the number
of evaluated subsets while searching for the best observation-subset models.
(g)
Let ρ̂j denote the minimal RSS for models selecting j observations. Furthermore, let ρ̂j denote its value
after g nodes of the regression tree have been generated (g = 0, . . . , 2n ). For any g, the RSS satisfy the
following relationship:
(g)
(g)
ρ̂1 ≤ ρ̂2 ≤ . . . ≤ ρ̂(g)
n .
See Lemma 1 in Appendix A for a formal proof. After the whole regression tree ∆(∅, I) has been generated, the minimal RSS corresponding to the best regression models for each number of observations are given
by ρ̂1 , . . . , ρ̂n (g = 2n ). Consider the gth node (S, A) and let its bound be ρS . A cutting test is devised.
Specifically,
if
(g−1)
ρS ≥ ρ̂nS +nA −i+1 , then
(g)
ρW ≥ ρ̂|W | ,
(3)
where W is any model obtained from ∆(S, Ai: ), i = 1, . . . , nA (see Lemma 2 for a formal proof). This implies
that the subtrees ∆(add(S, ai ), Ai+1: ) cannot improve ρ̂. A procedure to compute the regression tree follows.
The child nodes (add(S, ai ), Ai+1: ) of node (S, A) are computed from left to right, i.e. for increasing i. If the
bound ρS is greater than ρ̂nS +nA −i+1 , then the corresponding child and its younger siblings (i.e. child nodes to
the right) are discarded. Otherwise, the child node is computed and this procedure repeated for the next child.
This is illustrated in Algorithm 2. The break statement on line 11 terminates and exits the inner-most loop of
the algorithm. Note that the cutting test is not effective in the first p levels of the tree, where the RSS of the
submodels is 0.
The computational efficiency of the BBA improves when more nodes are cut. That is, if bigger subtrees are
bounded with bigger values. This can be achieved by preordering the observations in each node (S, A). The
BBA with preordering (PBBA) constructs nodes using “stronger” observations first. The observations in A are
sorted according to their strength (Agulló 2001). The exact bound RSS(S+ai ) can be computed to determine
the strength of observation ai ∈ A. Here, S+ai denotes the set S to which the observation ai has been added.
This approach involves nA rank-1 Cholesky updates (Gill et al. 1974, Golub & Van Loan 1996). Note that the
e denote the right-hand side of (2) and x
observations in A are sorted in decreasing order of the RSS. Let R
eT
the row vector corresponding to the added observation. The updating process is illustrated in Figure 3(a). It
8
Algorithm 2: Branch-and-bound algorithm (BBA)
1
procedure BBA(I, ρ̂, Ŝ)
2
n ← |I|
3
ρ̂i ← +∞, where i = 1, . . . , n
4
Insert node (∅, I) into node list
5
while list contains nodes do
6
Remove node (S, A) from node list
7
nS ← |S|; nA ← |A|; ρS ← RSS(S)
8
if ρS < ρ̂nS then ρ̂nS ← ρS , ŜnS ← S
9
for i = 1, . . . , nA do
10
if ρS ≥ ρ̂nS +nA −i+1 then break
11
Compute (S ′ , A′ ) = (add(S, ai ), Ai+1: )
12
Insert (S ′ , A′ ) into node list
13
end for
14
end while
15
end procedure BBA
involves the computation and the application of p + 1 Givens rotations, where p is the number of independent
variables. The ith Givens rotation Gi can be written as
ci si
,
Gi =
−si ci
2
ei,i /t, si = x
ei,i
where ci = R
ei /t and t2 = R
+x
e2i . The application of Gi is given by
(i)
(i−1)
e
e
Ri,i:p+1
Ri,i:p+1
Gi (i−1),T = (i),T ,
x
ei,i:p+1
x
ei,i:p+1
e(i) and x
e and x
e(0) ≡ R,
e x
where R
e(i),T respectively denote R
eT modified by the first i Givens rotations, R
e(0) ≡ x
e
and x
e(p+1) ≡ 0. The standard colon notation is used in order to denote submatrices and subvectors (Golub &
Van Loan 1996). The bound, i.e. the RSS, is given by the square of the element (p + 1, p + 1) of the matrix
e(p+1) = Gp+1 · · · G1 R.
e The procedure is computationally expensive. Note that the construction of a Givens
R
e Thus, the application of the sequence
rotation does not involve previously modified elements of the matrix R.
of Givens rotations can be carried out without explicitly modifying the upper triangular factor as illustrated in
(p)
2
ep+1,p+1
Figure 3(b). The bound is given by R
+ (e
xp+1 )2 . Thus, the computational complexity is roughly divided
by two. The new procedure to derive the bound is illustrated by Algorithm 3.
9
“
e (0)
R
x
e(0),T
”
G1
“
e (0)
R
x
e(0),T
”
G2
“
e (1)
R
x
e(1),T
”
G3
“
e (2)
R
x
e(2),T
”
G4
“
e (3)
R
x
e(3),T
”
G5
“
e (4)
R
x
e(4),T
”
“
e (5)
R
x
e(5),T
”
u u u u u
u u u u
u u u
u u
◦ u u u u
added
◦ u u u
zero
non-zero
◦ u u
u updated
u
◦
◦ u
◦ annihilated
⋆
⋆ bound
(a) Updating the QR decomposition after adding a row.
⋆
◦ u u u u
added
◦ u u u
zero
non-zero
◦ u u
◦ u
u updated
◦ annihilated
⋆
⋆ bound
(b) Efficiently computing the bound of the observation.
Figure 3: Exploiting the QR decomposition to compute the bound of an observation.
Algorithm 3: Computing the exact bound of an observation x
3
procedure bound(R, x, b)
for j = 1, . . . , p + 1 do
q
2
t ← Rj,j
+ x2j ; c ← Rj,j /t; s ← xj /t
4
5
6
7
xj ← −s · xj + c · Ri,j , where j = i + 1, . . . , p + 1
end for
b ← t2
end procedure
1
2
The absolute residuals (Resid) represent an alternative criterion that can be used in preordering the observations. In a node (S, A), the residual vector ε̂S is computed with respect to β̂S , the LS estimator of the
subset S of observations. This merely involves solving an upper-triangular system and is computationally non
expensive.
The traversal by the PBBA of the subsets-tree is divided into two stages. In stage I, the algorithm visits
the nodes that are on the first p levels (i.e. levels 0 to p − 1) of the regression tree. The remaining nodes
are visited in stage II. In stage I, the subset-models S are underdetermined, that is nS < p. The estimator β̂S
is not available, and neither the RSSs nor the residual vector can be computed. To circumvent this problem,
one can turn to another estimator, namely β̂S∪A . The initial linear model I is solved in the root node. Then,
in stage I of the algorithm, each new child node can be obtained by downdating the estimator of the parent
node by one observation. The downdating operation can be implemented, for example, by means of pseudoorthogonal rotations (Golub & Van Loan 1996, Alexander, Pan & Plemmons 1988). The residual vector ε̂S∪A
is then available and can be used to sort the available observations. Another indicator of the strength of an
observation is RSS(S ∪ A−ai ), the RSS of the estimator β̂S∪A downdated by the observation ai . This quantity
10
can be computed for every observation ai in A. The same scheme illustrated in Figure 3(b) can be employed,
substituting hyperbolic plane rotations for Givens rotations. The observations are then sorted in increasing
order of the criterion. To distinguish the two cases, RSS+ and Resid+ will denote the criteria used to preorder
the observations obtained by updating the estimator β̂S (stage II); RSS− and Resid− will denote the criteria
obtained by downdating β̂S∪A (stage I).
Preordering is an expensive procedure. The algorithm hence preorders the available observations in nodes
which are the root of big subtrees, i.e. where a potentially large number of nodes will be cut. A preordering
radius can be used to restrict the use of preordering (Hofmann et al. 2007). Specifically, observations are
preordered in nodes whose distance from the root node is smaller than the preordering radius. In the present
context this is equivalent to preordering the observations in all nodes such that nA > n − π, where π denotes
the preordering radius. Notice that if π = 0, then no preordering occurs. If π = 1, then the observations are
preordered in the root node; if π = n, then the observations are preordered in all nodes of the tree.
Figure 4 and 5 illustrate the computational cost of the PBBA for various preordering radii. The employed
dataset contains n = 40 observations (8 of which are outliers), p = 4 independent variables and is generated
according to Rousseeuw & Van Driessen (2006). The LTS estimates for coverage values hmin = 20 to hmax = 40
are computed. Four different preordering strategies are illustrated, depending on which criterion (RSS or Resid)
is used in each of the two stages (I/II). It can be seen from Figure 4 that all four preordering strategies generate
about the same number of nodes, and that the number of nodes does not continue to decrease for preordering
radii π > n/2. Thus, Resid should be the preordering criterion of choice as it is cheaper to compute than RSS,
and the employed preordering radius should be approximately n/2. These findings are confirmed by Figure 5
which shows that a preordering radius beyond n/2 does not significantly improve the execution time of the
algorithm. Moreover, employing the Resid criterion in stage II of the algorithm leads to execution times that
are up to 3 times lower for π ≥ n/2.
The PBBA is equivalent to Agulló’s (2001) algorithm (AGLA) under the conditions that follow. The AGLA
computes the LTS estimate for one coverage value only, i.e. h = hmin = hmax . Furthermore, preordering
of observations takes place in the root node with respect to the absolute residuals; in the second stage the
observations are sorted according to their RSS. In brief, the AGLA is equivalent to the PBBA with Resid−
(π1 = 1) / RSS+ (π2 = n), where π1 and π2 designate the preordering radii in stages I and II, respectively. For
a given range hmin : hmax the PBBA performs twice faster than the AGLA if it employs the same preordering
strategy. Contrary to the AGLA, the PBBA does not need to step through the regression tree more than once
and hence does not generate any redundant nodes.
Table 1 illustrates the results of an experimental comparison of the PBBA with the AGLA. The algorithms
are executed on datasets with n = 32, 36, 40, 44, 48 observations. The number of outliers is n/4. The PBBA
is executed with a preordering radius π = n/2. The LTS estimates are computed for three different ranges
11
hmin : hmax , namely n/2 : n, n/2 : 3n/4 and 3n/4 : n. The execution times and number of nodes indicated are
mean values taken over 100 different datasets generated for every data size. The experiment reveals that the
PBBA is 6 to 10 times faster than the AGLA. In certain cases, the AGLA generates less nodes then the PBBA.
It employs the RSS as a preordering criterion and preorders the observations in all nodes of stage II (i.e. down
to the leaf nodes). However, the RSS criterion is expensive to compute compared to Resid and a preordering
2.0e+07
1.5e+07
nodes
5.0e+06
1.0e+07
1.5e+07
1.0e+07
RSS− / RSS+
RSS− / Resid+
0
10
20
30
0.0e+00
0.0e+00
5.0e+06
nodes
2.0e+07
radius which is too large adds to the computational load. For these reasons the PBBA outperforms the AGLA.
40
Resid− / RSS+
Resid− / Resid+
0
10
preordering radius
20
30
40
preordering radius
(a) RSS− /Resid+ and RSS− /RSS+
(b) Resid− /Resid+ and Resid− /RSS+
Figure 4: Number of nodes visited by the PBBA with different preordering criteria in stages I/II of the algo-
40
30
10
20
execution time (sec)
30
20
10
execution time (sec)
40
rithm, as a function of the preordering radius (data: n = 40, p = 4).
0
Resid− / RSS+
Resid− / Resid+
0
0
RSS− / RSS+
RSS− / Resid+
10
20
30
40
0
10
preordering radius
20
30
40
preordering radius
(a) RSS− /Resid+ and RSS− /RSS+
(b) Resid− /Resid+ and Resid− /RSS+
Figure 5: Execution time (in seconds) of the PBBA with different preordering criteria in stages I/II of the
algorithm, as a function of the preordering radius (data: n = 40, p = 4).
12
Table 1: Time (in seconds) needed by the PBBA and AGLA to compute a range of LTS regressors, for three
different ranges hmin : hmax . The number of nodes visited by each algorithms is given in parentheses.
(a) hmin = n/2, hmax = n
n
p
q
32
36
40
44
48
3
3
4
4
5
8
9
10
11
12
32
36
40
44
48
3
3
4
4
5
8
9
10
11
12
AGLA
mean
min
Time in seconds
1.16
0.60
3.51
1.30
17.51
8.17
49.68
19.92
270.73
132.55
Number of nodes
99’434
50’322
268’115
91’892
974’200
462’648
2’506’074
987’432
10’477’284 5’146’928
max
mean
PBBA
min
max
2.71
11.08
52.44
123.88
1’164.31
0.18
0.54
2.34
6.75
31.58
0.092
0.16
0.91
2.56
12.16
0.40
1.52
6.74
16.36
122.47
6.44
6.50
7.48
7.36
8.57
238’444
881’282
2’964’662
6’292’358
44’844’933
97’138
275’415
1’061’924
2’966’393
12’276’869
44’742
72’665
424’824
1’021’971
4’724’249
215’763
821’995
3’168’084
7’517’632
46’391’496
1.02
0.97
0.92
0.84
0.85
AGLA / PBBA
(b) hmin = n/2, hmax = 3n/4
n
p
q
32
36
40
44
48
3
3
4
4
5
8
9
10
11
12
32
36
40
44
48
3
3
4
4
5
8
9
10
11
12
AGLA
mean
min
Time in seconds
0.87
0.47
2.47
0.92
13.43
7.52
36.03
17.16
205.12
114.84
Number of nodes
77’835
40’851
198’201
72’552
776’800
411’723
1’906’482
878’052
8’231’868 4’612’913
max
mean
PBBA
min
max
AGLA / PBBA
1.88
6.99
37.05
87.42
752.72
0.15
0.42
1.95
5.47
26.28
0.068
0.12
0.84
2.08
10.59
0.30
1.12
5.42
13.85
80.96
5.80
5.88
6.88
6.59
7.81
169’718
579’627
2’222’448
4’907’138
30’672’512
84’428
233’379
946’947
2’613’071
10’971’795
37’008
63’821
393’425
826’507
3’868’554
180’373
641’923
2’670’110
7’087’033
32’010’797
0.92
0.85
0.82
0.73
0.75
(c) hmin = 3n/4, hmax = n
n
p
q
32
36
40
44
48
3
3
4
4
5
8
9
10
11
12
32
36
40
44
48
3
3
4
4
5
8
9
10
11
12
AGLA
mean
min
Time in seconds
0.40
0.10
1.42
0.16
5.51
1.00
18.18
2.19
87.88
11.99
Number of nodes
29’901
6’560
96’467
9’662
271’261
47’830
811’691
88’112
3’044’579 362’517
max
mean
PBBA
min
max
AGLA / PBBA
1.44
5.83
25.71
60.85
753.21
0.061
0.21
0.67
2.13
8.79
0.012
0.02
0.11
0.24
1.18
0.22
0.76
2.99
6.88
64.96
6.67
6.76
8.22
8.54
10.00
114’792
432’003
1’353’596
2’857’928
27’933’149
21’804
72’002
194’495
580’998
2’139’972
3’964
5’609
28’420
53’587
209’355
100’873
336’567
1’093’940
2’248’152
20’633’518
1.37
1.34
1.40
1.40
1.42
13
4
Conclusions
Various strategies to compute the exact least-trimmed-squares (LTS) regression are proposed. The adding
row algorithm (ARA) is based on a regression tree. The computational tool employed is a rank-1 Cholesky
updating procedure. The exploitation of the tree’s structure allows to determine LTS estimates for a range
of coverages. Thus, the coverage parameter h does not need to be known in advance and the algorithm can
be used to examine the degree of contamination of the data. The branch-and-bound algorithm (BBA) avoids
the explicit enumeration of all observation-subset models. It considerably reduces the execution time. The
BBA with observation preordering (PBBA) sorts the observations in each node to increase the computational
performance of the BBA. Experiments show a significant improvement of the PBBA over the BBA. In the ARA
context a fast computation of the bounds has been devised.
A heuristic strategy which provides solutions reasonably close to the best solution can lead to smaller execution times. The heuristic BBA (HBBA) uses a tolerance parameter τ to cut subtrees (Gatu & Kontoghiorghes
2006). Although very efficient for variable-subset regression, this approach does not show interesting results
when combined with the PBBA. The increase in computational efficiency with respect to the loss in quality of
the solution computed by the HBBA, is not significant. The PBBA traverses the regression tree starting from
the empty model, updating submodels as it procedes downward. Thus, τ has little effect on the execution time
becuase the residual sum of squares (RSSs) are small in early stages of the algorithm.
The PBBA is compared to the exact algorithm (AGLA) presented by Agulló (2001). The AGLA computes
the LTS regressor for a given coverage parameter h, usually n/2. Thus, it will discard relevant data as the
degree of contamination of the data is typically less than 50% and might fail to reveal the global linear trend
of the data. The PBBA can be seen as a generalization of the AGLA. It uses a more general notion of the
subsets-tree to compute a set of LTS regressors for a range of coverage values hmin : hmax simultaneously.
Furthermore, the PBBA employs a more efficient preordering strategy resulting in a smaller computational
load. Experiments show that the PBBA is 6 to 10 times faster than the AGLA to compute a range of LTS
regressors. It is an efficient tool to examine the degree of contamination of the data, hence revealing the exact
LTS estimator which is both robust and accurate.
Acknowldegements
The authors are grateful to the two anonymous referees for their valuable comments and suggestions.
14
A
Formal proofs
The proofs of the Lemmas 1–3 are given. These follow closely the proofs given in (Gatu & Kontoghiorghes
2006).
(g)
(g)
Lemma 1 rj ≤ rj+1 (j = 1, . . . , n − 1).
(0)
Proof. 1 The proof is by induction on the number of generated nodes. Initially, rj = +∞ and the proposition
holds. By inductive hypothesis, the proposition holds if g nodes have been generated. It must be shown that the
proposition holds after the (g + 1)th node has been computed.
Consider the (g + 1)th node (S, A), with nS = j. It selects the observations in S and affects rj which
(g+1)
when modified becomes rj
(g+1)
(g)
= RSS(S) ≤ rj . Thus, by inductive hypothesis, rj
(g)
(g+1)
≤ rj+1 = rj+1 . The
model S was derived from its parent model Spar by adding an observation. Hence, RSS(S) ≥ RSS(Spar ) and
(g+1)
(g+1)
rj−1 ≤ RSS(Spar ) ≤ rj
. This completes the proof.
Lemma 2 Given the node (S, A) and a constant α > 0,
if
(g)
rnS +nA ≤ α · RSS(S) then
(g)
r|W | ≤ α · RSS(W ),
where W is any observation-subset obtained from ∆(S, k).
Proof. 2 Any observation-subset W of size j (j = nS + 1, . . . , nS + nA ) was obtained by adding one or
(g)
more observations to S. Thus, RSS(W ) ≥ RSS(S). From Lemma 1 follows that rj
(g)
(g)
≤ rnS +nA . Hence, if
(g)
rnS +nA ≤ α · RSS(S) then rj ≤ α · RSS(W ). This completes the proof.
Lemma 3 Let Se be an observation-subset model selected by the HBBAτ . Then,
e ≤ τ.
RRE(S)
e =
Proof. 3 Two possibilities arise. Either the HBBAτ found the optimal model, i.e. Se = S ∗ , and RRE(S)
e = |S ∗ | = nS . According to
0 ≤ τ . Else, the model S ∗ was contained in a subtree that was cut. Note that |S|
(g)
e ≤ (1+τ )·RSS(S ∗ )
e ≤ r(g) . Thus, RSS(S)
Lemma 2 with α = 1+τ , rnS ≤ (1+τ )·RSS(S ∗ ). Further, RSS(S)
j
which completes the proof.
References
Agulló, J. (2001), ‘New algorithms for computing the least trimmed squares regression estimator’, Computational Statistics and Data Analysis 36, 425–439.
15
Alexander, S. T., Pan, C.-T. & Plemmons, R. J. (1988), ‘Analysis of a recursive least squares hyperbolic rotation
algorithm for signal processing’, Linear Algebra and its Applications 98, 3–40.
Atkinson, A. C. & Cheng, T.-C. (1999), ‘Computing least trimmed squares regression with the forward search’,
Statistics and Computing 9, 251–263.
Belsley, D. A., Kuh, A. E. & Welsch, R. E. (1980), Regression Diagnostics: Identifying Influential Data and
Sources of Collinearity, John Wiley and Sons, New York.
Björck, Å., Park, H. & Eldén, L. (1994), ‘Accurate downdating of least squares solutions’, SIAM Journal on
Matrix Analysis and Applications 15(2), 549–568.
Burks, A. W., Warren, D. W. & Wright, J. B. (1954), ‘An analysis of a logical machine using parenthesis-free
notation’, Mathematical Tables and Other Aids to Computation 8(46), 53–57.
Dongarra, J. J., Bunch, J. R., Moler, C. B. & Stewart, G. W. (1979), LINPACK Users’ Guide, SIAM, Philadelphia.
Donoho, D. L. & Huber, P. J. (1983), The notion of breakdown point, in P. J. Bickel, K. A. Doksum, E. L.
Lehman & J. L. Hodges, eds, ‘A festschrift for Erich L. Lehmann in honor of his sixty-fifth birthday’,
CRC Press.
Furnival, G. & Wilson, R. (1974), ‘Regression by leaps and bounds’, Technometrics 16, 499–511.
Gatu, C. & Kontoghiorghes, E. J. (2003), ‘Parallel algorithms for computing all possible subset regression
models using the QR decomposition’, Parallel Computing 29(4), 505–521.
Gatu, C. & Kontoghiorghes, E. J. (2006), ‘Branch-and-bound algorithms for computing the best subset regression models’, Journal of Computational and Graphical Statistics 15, 139–156.
Gill, P. E., Golub, G. H., Murray, W. & Saunders, M. A. (1974), ‘Methods for modifying matrix factorizations’,
Mathematics of Computations 28, 505–535.
Golub, G. H. & Van Loan, C. F. (1996), Matrix computations, Johns Hopkins Studies in the Mathematical
Sciences, 3rd edn, Johns Hopkins University Press, Baltimore, Maryland.
Hawkins, D. M. (1994), ‘The feasible solution algorithm for least trimmed squares regression’, Computational
Statistics and Data Analysis 17, 185–196.
Hawkins, D. M. & Olive, D. J. (1999), ‘Improved feasible solution algorithms for high breakdown estimation’,
Computational Statistics and Data Analysis 30, 1–11.
16
Hofmann, M., Gatu, C. & Kontoghiorghes, E. J. (2007), ‘Efficient algorithms for computing the best-subset
regression models for large-scale problems’, Computational Statistics and Data Analysis 52, 16–29.
Hössjer, O. (1994), ‘Rank-based estimates in the linear model with high breakdown point’, Journal of the
American Statistical Association 89, 149–158.
Morgenthaler, S. (1991), ‘A note on efficient regression estimators with positive breakdown point’, Statistics
and Probability Letters 11, 469–472.
Narendra, P. M. & Fukunaga, K. (1977), ‘A branch and bound algorithm for feature subset selection’, IEEE
Transactions on Computers 26(9), 917–922.
Newell, A. & Shaw, J. C. (1957), Programming the logic theory machine, in ‘Proceedings of the Western Joint
Computer Conference’, Institute of Radio Engineers, pp. 230–240.
Rousseeuw, P. J. (1984), ‘Least median of squares regression’, Journal of the American Statistical Association
79, 871–880.
Rousseeuw, P. J. (1997), Introduction to positive-breakdown methods, in G. S. Maddala & C. R. Rao, eds,
‘Handbook of statistics’, Vol. 15: Robust inference, Elsevier, pp. 101–121.
Rousseeuw, P. J. & Leroy, A. M. (1987), Robust regression and outlier detection, John Wiley & Sons.
Rousseeuw, P. J. & Van Driessen, K. (2006), ‘Computing LTS regression for large data sets’, Data Mining and
Knowledge Discovery 12, 29–45.
Smith, D. M. & Bremner, J. M. (1989), ‘All possible subset regressions using the QR decomposition’, Computational Statistics and Data Analysis 7(3), 217–235.
Stefanski, L. A. (1991), ‘A note on high-breakdown estimators’, Statistics and Probability Letters 11, 353–358.
17
Matrix strategies for computing the least trimmed squares
estimation of the general linear and SUR models
Marc Hofmann∗,
Erricos John Kontoghiorghes†
Abstract
An algorithm has been recently proposed for computing the exact least trimmed squares (LTS)
estimator of the standard regression model. This combinatorial algorithm is adapted to the case
of the general linear and seemingly unrelated regression models with possible singular dispersion
matrices. It searches through a regression tree to find optimal estimates by employing efficient
matrix techniques to update the generalized linear least squares problem in each tree node. The
new formulation of the problem allows one to update the residual sum of squares of a subset model
efficiently. Specifically, the new algorithms utilize previous computations in order to update the
generalized QR decomposition by a single observation. The sparse structure of the models are
exploited. Theoretical measures of computational complexity are provided. Experimental results
confirm the ability of the algorithms to identify the outlying observations, and at the same time,
they illustrate the computational intensity of deriving the LTS estimators.
Keywords: Least trimmed squards, general linear model, seemingly unrelated regressions, generalized
linear least squares.
1 Introduction
Algorithms for least trimmed squares (LTS) regression of the ordinary linear model have been proposed
(Agulló 2001, Rousseeuw & Leroy 1987, Rousseeuw & Van Driessen 2006). Robust multivariate
methods, and multivariate LTS in particular, have also been investigated (Agulló, Croux & Aelst 2008,
∗
†
Institut d’informatique, Université de Neuchâtel, Switzerland. E-mail: marc.hofmann@unine.ch
Department of Public and Business Administration, University of Cyprus, Cyprus; and School of Computer Science
and Information Systems, Birkbeck College, University of London, UK.
1
E-mail: erricos@ucy.ac.cy
Hubert, Rousseeuw & Aelst 2008, Rousseeuw, Aelst, Driessen & Agulló 2004). Here, new numerical
strategies to solve the LTS regression of the general linear model (GLM) and of the seemingly unrelated
regressions (SUR) model are designed. These strategies exploit the matrix properties of the linear
models. Recently, a fast branch and bound strategy for computing the LTS estimator has been designed
(Hofmann, Gatu & Kontoghiorghes 2008). This algorithm is extended to compute the LTS estimator of
the GLM and SUR model (Srivastava & Giles 1987, Srivastava & Dwivedi 1979, Zellner 1962). The
GLM is given by:
y = Xβ + ε,
(1)
where y ∈ Rm , X ∈ Rm×n , β ∈ Rn and ε ∼ (0, σ 2 Ω), Ω ∈ Rm×m (m ≥ n). The objective of the LTS
P
estimator is to minimize hi=1 e2(i) , where e2(1) , . . ., e2(m) are the squared residuals sorted in increasing
order and h is the coverage parameter (h ≥ ⌊m + n + 1⌋/2). This is equivalent to finding the h-subset
with the smallest least-squares (LS) objective function.
The SUR model is a special case of the GLM and is written as:
y (i) = X (i) β (i) + ε(i) ,
i = 1, . . . , G,
(i)
where y (i) ∈ Rm , X (i) ∈ Rm×ni (m ≥ ni ), β (i) ∈ Rni and ε(i) ∈ Rm . Furthermore, E(εk ) = 0, and
(i)
(j)
(i)
(j)
contemporaneous disturbances are correlated, i.e. Var(εt , εt ) = σij and Var(εs , εt ) = 0 if s 6= t.
In compact form, the model may be written:
G
vec(Y ) = ⊕ X (i) · vec({β (i) }G ) + vec(E),
i=1
where Y = y (1) · · · y (G)
∈ Rm×G , E = ε(1) · · · ε(G)
(2)
∈ Rm×G , vec(E) ∼ (0, Σ ⊗ Im )
(i)
and Σ = (σij ) ∈ RG×G . The set of vectors β (1) , . . . , β (G) is denoted by {β (i) }G , ⊕G
=
i=1 X
diag(X (1) , . . . , X (G) ) denotes the direct sum of the matrices X (1) , . . .,X (G) and vec(•) is the vector
operator which stacks a set of column vectors.
Here the GLM and SUR model are reformulated as a generalized linear least squares problem
(GLLSP) (Foschi & Kontoghiorghes 2002, Kontoghiorghes & Clarke 1995, Paige 1978, Paige 1979b,
Paige 1979a). The solution of the GLLSP using orthogonal factorization methods is numerically stable.
Furthermore, the GLLSP can be updated efficiently after one observation has been added to the regression model (Kontoghiorghes 2004, Yanev & Kontoghiorghes 2007). The best linear unbiased estimator
(BLUE) of β in (1) is the solution of the GLLSP
βb = argmin kuk2
β
subject to
2
y = Xβ + Bu,
(3)
where B ∈ Rm×p such that Ω = BB T , u ∼ (0, σ 2 Im ) and p ≥ m − n. The residual sum of squares of
b = kuk2 . Notice that B is not necessarily of full rank.
βb is RSS(β)
Similarly, the BLUE of β in the SUR model (2) is obtained by solving the GLLSP (hereafter SURGLLSP):
{βb(i) }G = argmin kY kF
subject to
{β (i) }G
G
vec(Y ) = ⊕ X (i) · vec({β (i) }G ) + K · vec(U ),
i=1
(4)
where k • kF denotes the Frobenius norm, E = U C T , C ∈ RG×G is upper triangular such that CC T =
Σ, K = C ⊗ Im , vec(U ) ∼ (0, σ 2 IM ) and M = Gm. The residual sum of squares is given by
RSS({βb(i) }G ) = kU kF .
Section 2 provides a brief description of the row-adding algorithm which computes the LTS estimators for a range of coverage values. Sections 3 and 4 present the adaptation of the LTS algorithm
for the GLM and SUR model, respectively. Within this context, emphasis is given to the development
of efficient strategies for updating the linear models. The sparse matrix structures are exploited for
minimizing the cost of expensive matrix operations. Experimental results are also presented. Section 5
concludes.
2 The row-adding algorithm
The row-adding algorithm (ARA) generates all possible observation-subset models to compute the
LTS estimates for a range hmin , . . . , hmax of coverage values (Hofmann et al. 2008). The observation
subsets are organized in a regression tree illustrated in Figure 1 (Belsley, Kuh & Welsch 1980, Gatu
& Kontoghiorghes 2003). A node (S,A) corresponds to a set of selected observations S and a set of
available observations A. The observations in A are used to compute new nodes. For example, in
the node ([12],[34]) on level 2, the observations 1 and 2 are selected and new nodes can be computed
by adding observations 3 or 4. If observation 4 is selected, then 3 is skipped so as to avoid generating
duplicate subsets. Thus, it is ensured that all submodels are generated and that no submodel is computed
more than once. A branch and bound strategy is employed to reduce computational time (Furnival &
Wilson 1974, Gatu & Kontoghiorghes 2006, Hofmann et al. 2008). The RSS is computed in each node
by updating the numerical quantities computed in the parent node. Effectively, this corresponds to
updating a generalized QR decomposition after a row has been added. The best RSS for every subset
size is stored in a list, which is updated each time a better solution for one of the subset sizes is found.
This is illustrated in Algorithm 1. The input argument I is the list of observation indices contained in
3
the data. The output arguments are the RSS and the indices of the selected observations (SEL) of the
computed LTS estimators.
[], [1234]
level 0
[1], [234]
1
2
[12], [34]
3
[123], [4]
4
[1234], []
[13], [4]
[124], []
[2], [34]
[14], []
[134], []
[23], [4]
[24], []
[234], []
Figure 1: The ARA regression tree for m = 4 observations.
Algorithm 1: Adding row algorithm (ARA)
1
procedure ARA(I, OPT, SEL)
2
m ← |I|
3
OPT(i) ← +∞, where i = 1, . . . , m
4
Insert node (∅, I) into node list
5
while list contains nodes do
6
Remove node (S, A) from node list
7
if RSS(S) <OPT(|S|) then OPT(|S|) ←RSS(S), SEL(|S|) ← S
8
for i = 1, . . . , |A| do
9
if RSS(S) ≥OPT(|S| + |A| − i + 1) then break
10
Compute (S ′ , A′ ) = add(S, A, i)
11
Insert (S ′ , A′ ) into node list
12
end for
13
end while
14
end procedure ARA
4
[3], [4]
[34], []
[4], []
3 The LTS estimation of the GLM
For the solution of the GLLSP (3), the generalized QR decomposition (GQRD) of X and B is required:
!
!
R
T
T
11
11
12
QT X =
and QT BP T =
.
(5)
0
0 T22
Here Q ∈ Rm×m and P ∈ Rp×p are orthogonal, T11 ∈ Rn×(p−m+n) is upper trapezoidal, T12 ∈
Rn×(m−n) and T22 ∈ R(m−n)×(m−n) is upper triangular. The constraints y = Xβ +Bu of the GLLSP (3)
are equivalent to QT y = QT Xβ + QT BP T P u. Thus, the transformed GLLSP can be written as:
!
!
!
!
y
R
T
T
u
1
11
11
12
1
βb = argmin ku1 k2 + ku2 k2 subject to
=
β+
,
β
y2
0
0 T22
u2
y1
!
u1
(6)
!
and P u =
. Thus, y2 = T22 u2 and the solution z = R11 βb follows from
y2
u2
u1 = 0, where z = y1 − T12 u2 . The RSS is ku2 k2 . Notice that (6) is equivalent to
where Q y =
T
βb = argmin ku1 k2
β
subject to z = R11 β + T11 u1 .
(7)
b
This is the reduced-size GLLSP of (1) and has the same solution β.
It is not necessary to compute the new solution from scratch after an observation has been added to
the GLM. Let the updated GLM (1) be given by:
e + εe,
ye = Xβ
where
ye =
υ
!
y
e=
X
,
xT
(8)
!
e B ),
and εe ∼ (0, σ B
2
X
eT
with
bT
e=
B
B
!
.
The GLLSP corresponding to (8) is:
βe = argmin ku1 k2
β
ζ
subject to
z
!
=
xT
R11
!
β+
!
b1 T
T11
u1 ,
(9)
where bT P T = b1 T b2 T , ζ = υ − b2 T u2 , u2 is given by (6) and z, R11 , T11 by (7). Now consider the
updated GQRD
eT
Q
xT
R11
!
=
e11
R
0
!
eT
Q
and
5
b1 T
T11
!
PeT =
Te11 t
0
τ
!
.
(10)
After applying the orthogonal transformations to (9), the GLLSP becomes:
!
!
!
!
e
e
y
e
R
T
t
u
e
1
11
11
1
βe = argmin ke
u1 k2 + η 2 subject to
=
β+
,
β
0 τ
0
η
ξ
where
eT
Q
ζ
!
z
=
ye1
ξ
!
and
Peu1 =
u
e1
η
(11)
!
.
e where ze = ye1 − tη. The new RSS is
e11 β,
Hence, η = ξ/τ and setting u
e1 = 0 leads to ze = R
e = RSS(β)
b + η 2 . As expected, the RSS is non-decreasing an observation is added to the GLM.
RSS(β)
The reduced GLLSP of (8) is given by:
βe = argmin ke
u1 k2
β
e11 β + Te11 u
subject to ze = R
e1 .
(12)
It follows that if the row υ xT bT has been properly transformed — or reduced — to ζ xT bT1
in (9), then the solution of the updated GLM (8) can be obtained from the reduced GLLSP (7). This
is exploited by the ARA to solve all subset models. The ARA solves a new subset model S ′ in each
node (S ′ , A′ ) by updating the model S of the parent node (S, A) with one of the observations in A.
Specifically, given a GLLSP (7) it computes the updated GQRD (10) in order to obtain the reduced
GLLSP (12) of the new subset model. All observations in A′ are reduced so that they can be used later
in updating the new, reduced GLLSP.
The GLM is underdetermined on the first n levels of the tree (i.e. m < n). On level n, the ARA
computes the GQRD (5) and it transforms the GLLSP according to (6) with m = n. The matrix
structure of the GLLSP is illustrated in Figure 2(a). Both sets of observations are shown: the set S of n
selected points (y, X, B) and the set A of k available points (yA , XA , BA ). As illustrated in Figure 2(b),
the orthogonal transformation P T must be applied to BA .
Following the order imposed by the all subsets tree, each of the k observations in A are selected in
turn to compute a new subset model. As illustrated in Figure 3(a), the ith available observation is added
to the GLLSP, while the first i − 1 are discarded. In Figure 3(b), the GLLSP is transformed according
to (11). Finally, the reduced GLLSP (12) is formed in Figure 3(c).
The GLLSP (11) is computed by updating the GQRD using Givens rotations (Golub & Van Loan
1996, Paige 1978) as illustrated in Figure 4. In Figure 4(a), a row is added to (7) to form (9). In
Figure 4(b), (p − n − 1) Givens rotations are applied to the right of T11 and BA . Next, four pairs of
ei from the left annihilates one subdiagonal
Givens rotations are applied in Figure 4(c)-4(f). A rotation Q
element in R11 , causing the fillup of a subdiagonal element in T11 . The fillup is annihilated by a Givens
rotation Pej applied from the right. Finally, the GLLSP is reduced in Figure 4(g).
6
k yA
XA
BA
k
n y
X
B
n
n
p
n
(a) Structure of the
(b)
GLLSP.
GLLSP.
zero
non-zero
The
p
transformed
modified element
Figure 2: Computing the GQRD and transforming the GLLSP (case p > n).
k−i
k−i
k−i
1
n
n+1
n
n
p
n
(a) Add row.
p
1
(b) Update GQRD.
zero
non-zero
added
n
p−1
1
(c) Reduce GQRD.
modified element
Figure 3: Updating the GLLSP with the i-th observation (case p > n).
Consider the GLLSP (11) with dimensions n and p. Let T (n, k, p) denote the cost of computing all
possible subset models, where k is the number of available observations (n ≥ 1, p ≥ k ≥ 0). It can be
expressed as:
T (n, k, p) =
k
X
[Tu (n, k − i, p) + T (n, k − i, p − 1)]
i=1
= Tu (n, k, p) + T (n, k − 1, p − 1) + T (n, k − 1, p),
where Tu is the cost of the updating operation. The closed form for T is given by:
p k X
X
k−i
T (n, k, p) =
Tu (n, i, j).
p−j
i=0 j=i
The complexity Tu (n, k, p) expressed in terms of elementary rotations is:
3n2 /2 + (k + 1)p + 4n − (k + 1)/2
Tu (n, k, p) =
n2 /2 + 2np − p2 + n + (k + 4)p − (k + 5)/2
7
if
p > n,
if
p ≤ n.
•
•
k
•
•
1×
•
•
n
•
•
•• ••
•• ••
•• ••
•• ••
××××
•• ••
• ••
••
•
n
•• •• •• ••
•• •• •• ••
•• •• •• ••
•• •• •• ••
××××××××
•• ••
• ••
••
•
•
•
•
•
•
•
•
•
•
p
•
•
•
•
•
•
•
•
•
•
•
◦
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
• •
• •
• •
• •
• •
◦ •
×
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
◦
•
•
•
•
•
•
•
•
e T , PeT .
(c) Q
1
4
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
◦
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
◦
•
•
•
•
◦
•
•
•
•
◦
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
• •
• •
• •
• •
• •
• •
◦ •
×
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
e T , PeT .
(d) Q
2
5
•
•
•
•
•
•
•
•
•
•
•
• •
• •
• •
• •
• •
• •
• •
◦ •
×
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
e T , PeT .
(e) Q
6
3
•
•
k
•
•
•
•
n
•
•
•
•
•
•
•
•
•
•
•
•
(b) Pe1T , Pe2T , Pe3T .
(a) Add row.
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
◦
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
••
••
••
••
••
••
••
••
◦•
×
e T , PeT .
(f) Q
7
4
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
n
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
p−1
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
(g) Reduce GQRD.
zero
• non-zero
◦ zeroed
× fillup
modified element
8
Figure 4: Givens sequence for updating the GLM (case p > n).
In computing the complexity Tu , it is assumed that the cost of adding a multiple of a vector to another
vector of the same length (Figure 3(c)) is equal to half the cost of rotating the same vectors. Furthermore, assuming that the cost of solving the GQRD (Figure 2) is equal to the cost of n update operations,
it follows that the overall cost of the ARA is T (n, m, m) ∈ O (2m (m2 + n2 )) when p = m.
3.1 Experimental results
The GLM is simulated with m observations and n variables. The chosen variance structure is given by:
Ωi,j =
σ2
ϕ|i−j| .
1 − ϕ2
(13)
The values of σ = 0.25 and ϕ = 0.9 were chosen for the simulation. Specifically,
y = Xβ + Bu,
where β, u ∼ (0, 1), X ∼ (0, 10) and B is the upper triangular Cholesky factor of the variancecovariance matrix Ω. Outliers are introduced by replacing some values of y by values that are normally
distributed with mean 100 and standard deviation 10. Five instances are simulated for each of the
different dimensions of the problem. The mean execution times are shown in Table 1. The experiments show that the computational cost remains high and the algorithm cannot be used for large-scale
problems. The algorithm correctly identified the outlying observations in all simulations. Figure 5
illustrates the RSSs computed by the algorithm for the simulated GLM with m = 24 observations. The
steep increase in RSS for subset sizes greater than 18 indicates that the computed LTS estimators for a
coverage h > 18 are contaminated by outliers.
Table 1: Mean execution times in seconds of the LTS algorithm for simulated GLM models.
m
n
outliers
16
4
4
4
20
4
5
14
24
4
6
62
28
8
7
4’404
32
8
8
18’038
36
8
9
62’551
9
time (s)
8e+05
RSS
4e+05
0e+00
5
10
15
20
size
Figure 5: The LTS-RSSs computed for the simulated GLM with m = 24 observations.
4 The LTS estimation of the SUR model
For the solution of the SUR-GLLSP (4), which provides the estimator of the SUR model (2), consider
the GQRD of ⊕ X (i) and K = C ⊗ Im :
QT · ⊕ X (i) =
⊕ R(i)
!
and
0
T11 T12
QT KP T =
0
T22
!
,
where R(i) ∈ Rni ×ni , T11 ∈ RN ×N , T22 ∈ R(M −N )×(M −N ) are upper-triangular and Q ∈ RM ×M is
(i)
orthogonal. Here, Q = ⊕ Q(i)
⊕ Q2 , where the QRD of X (i) is given by
1
(i) T
Q
X (i) =
R(i)
!
0
, with
(i)
Q(i) = Q(i)
Q
1
2
such that
(i) T
Q1 X (i) = R(i)
The SUR-GLLSP becomes
b(i)
2
2
{β } = argmin(ku1 k + ku2 k ) subject to
{β (i) }
where
y1
y2
!
y1
y2
(i) T
and Q2 X (i) = 0.
!
=
⊕ R(i)
(i) T
⊕ Q1
· vec(Y ) and
=
(i) T
⊕ Q2
!
0
u1
u2
(i)
· vec({β }) +
0
T22
!
u1
u2
!
,
!
= P · vec(U ).
Thus, u1 = 0 and y2 = T22 · u2 . The estimator of the SUR model is given by:
⊕ R(i) · vec({βb(i) }) = vec({z (i) }),
10
T11 T12
(14)
where vec({z (i) }) = y1 − T12 · u2 , z (i) ∈ Rni and RSS({βb(i) }) = ku2 k2 . This is equivalent to
solving the triangular systems R(i) βb(i) = z (i) (i = 1, . . . , G). The procedure is illustrated in Figure 6.
An orthogonal transformation from the left is applied to triangularize the X (i) s. Next, the rows are
permuted such as to obtain ⊕ R(i) in the upper part of the data matrix. The same permutation is applied
to the rows and columns of the covariance structure K. An orthogonal transformation is applied to the
right of K to form a block upper-triangular structure. Finally, the GLLSP is reduced by a block column
to:
{βb(i) } = argmin ku1 k2
subject to
{β (i) }
vec({z (i) }) = ⊕ R(i) · vec({β (i) }) + T11 · u1 .
An observation is added to each of the G equations of the SUR model (2). The reduced SURGLLSP is now written as:
b(i)
2
2
{β } = argmin(ku1 k +ku2 k ) subject to
{β (i) }
vec({z (i) })
!
=
vec({ζ (i) })
⊕ R(i)
⊕ a(i)
!
(i)
·vec({β })+
T
T11
0
0
C
!
u1
!
u2
.
The GQRD of ⊕ R(i) and T11 is updated by a sequence of Givens rotations to give:
!
!
!
!
(i)
e11 Te12
e(i)
⊕
R
T
⊕
R
T
0
11
eT
eT
Q
=
PeT =
and Q
.
T
⊕ a(i)
0
0 Te22
0 C
Thus, the SUR-GLLSP is reduced to:
{βe(i) } = argmin(kv1 k2 +kv2 k2 ) subject to
{β (i) }
where
ye1
!
vec({ξ (i) })
eT
=Q
!
ye1
vec({ξ (i) })
vec({z (i) })
=
!
vec({ζ (i) })
and
e(i)
⊕R
!
0
v1
v2
!
Te11 Te12
·vec({β (i) })+
0 Te22
u1
= Pe
u2
!
v1
v2
!
.
e(i) · vec({βe(i) }), where vec({e
The solution is given by vec({e
z (i) }) = ⊕ R
z (i) }) = ye1 − Te12 · v2 . The
RSS of the new model is RSS({βe(i) }) = RSS({βb(i) }) + kv2 k2 . The various steps of the procedure are
illustrated schematically in Figure 7. The new observations are appended to the data matrix ⊕ R(i) and
the block C to the covariance structure T11 . The G equations are updated by applying the orthogonal
eT from the left. Next, the block upper-triangular structure of T11 is restored by an ortransformation Q
thogonal transformation from the right. Finally, the model is reduced. Givens sequences are employed
to update the matrices (Foschi, Belsley & Kontoghiorghes 2003, Kontoghiorghes 2004).
The ARA can be modified to compute the all-observation-subsets regression for the SUR model. A
remarkable difference with the GLM is that observations that are available for selection are not affected
11
!
,
vec(Y )
⊕ X (i)
K
(a) The SUR-GLLSP.
(b) Apply QT from the left.
(c) Permute rows and columns.
(d) Apply P T from the right.
vec({z (i) })
⊕ R(i)
T11
(e) Reduce SUR-GLLSP.
zero
non-zero
modified
element
Figure 6: Solving the SUR-GLLSP (G = 3).
by the orthogonal transformation P T . In constructing the model, the G regressions are sorted in order
of the increasing numbers of variables. Hence, nG ≥ ni , i = 1, . . . , G. A subset model is solved in
each node of level nG as illustrated in Figure 6. The algorithm proceeds by updating the GQRD with
one observation at a time (Figure 7).
12
vec({z (i) })
⊕ R(i)
T11
C
(b) Apply QT from the left.
(a) Add rows.
vec({e
z (i) })
(c) Apply P T from the right.
zero
non-zero
e(i)
⊕R
Te11
(d) Reduce SUR-GQRD.
added
modified
element
Figure 7: Updating the SUR-GLLSP (G = 3).
The cost of computing the subtree of a node residing in a level ℓ > nG is:
T (G, {ni }, k) =
k
X
[Tu (G, {ni }) + T (G, {ni }, k − i)]
i=1
= 2k+1 − 1 · Tu (G, {ni }),
where Tu is the cost of updating the SUR-GQRD, Tu (G, {ni }) ∈ O(GN 2 ) and N =
P
ni . Assuming
that solving the GQRD in level nG has the same cost as performing nG updates, the complexity of the
algorithm is T (G, {ni }, m) ∈ O(2m GN 2 ).
4.1 Experimental results
The SUR model is simulated as follows:
vec({y (i) }) = ⊕ X (i) · β + (C ⊗ Im ) · u,
13
where X (i) ∼ (0, 10) and β, u ∼ (0, 1), C is the upper-triangular Cholesky factor of Σ, which is
generated according to the same scheme as Ω in (13). Outlying observations are simulated by replacing
some values at identical indices in each of the vectors y (i) by values that are normally distributed with
mean 100 and standard deviation 10. Five instances of the problem are simulated for each of various
dimensions. The mean execution times are illustrated in Table 2. The algorithm is computationally
infeasible for large-scale SUR models. Figure 8 reveals, as was the case for the GLM, the degree of
contamination of the simulated data.
Table 2: Mean execution times in seconds for the simulated SUR model.
m
ni (G = 3)
outliers
16
4,6,8
4
12
20
4,6,8
5
96
24
4,6,8
6
537
28
6,8,10
7
10561
32
6,8,10
8
46120
0
RSS
5000 10000
20000
time (s)
5
10
15
20
size
Figure 8: The LTS-RSSs computed for the simulated SUR with m = 24 observations.
5 Conclusions
New algorithms have been designed to compute the LTS estimators of the GLM and SUR model. These
algorithms are based on LTS strategies for the standard regression model. The singularity problem of
14
the dispersion matrix is avoided by reformulating the estimation problem as a GLLSP. The main computational tool of the algorithm is the GQRD. Efficient strategies to update the GQRD are employed.
The sparse matrix structure of the linear models are exploited to apply orthogonal transformations efficiently. Thus, solutions to the GLLSP are obtained efficiently and without having to solve the GLLSP
from scratch.
Although the algorithm is infeasible for larger problem sizes, simulations that it succeeds in detecting outlying observations. Future work can address the LTS problem of the SUR model where the
regressions are modified independently. That is, observations are deleted from one regression at a time.
This will yield an unbalance SUR model in each node of the ARA tree (Foschi & Kontoghiorghes
2002). The feasability of adapting the Fast LTS algorithm (Rousseeuw & Van Driessen 2006) to the
GLM and the use of parallel computing in order to solve larger scale problems merits investigation.
References
Agulló, J. (2001), ‘New algorithms for computing the least trimmed squares regression estimator’,
Computational Statistics and Data Analysis 36, 425–439.
Agulló, J., Croux, C. & Aelst, S. V. (2008), ‘The multivariate least-trimmed squares estimator’, Journal
of multivariate analysis 99(3), 311–338.
Belsley, D. A., Kuh, A. E. & Welsch, R. E. (1980), Regression Diagnostics: Identifying Influential Data
and Sources of Collinearity, John Wiley and Sons, New York.
Foschi, P., Belsley, D. A. & Kontoghiorghes, E. J. (2003), ‘A comparative study of algorithms for
solving seemingly unrelated regressions models’, Computational Statistics and Data Analysis
44(1-2), 3–35.
Foschi, P. & Kontoghiorghes, E. J. (2002), ‘Seemingly unrelated regression model with unequal size
observations: computational aspects’, Computational Statistics and Data Analysis 41(1), 211–
229.
Furnival, G. & Wilson, R. (1974), ‘Regression by leaps and bounds’, Technometrics 16, 499–511.
Gatu, C. & Kontoghiorghes, E. J. (2003), ‘Parallel algorithms for computing all possible subset regression models using the QR decomposition’, Parallel Computing 29(4), 505–521.
15
Gatu, C. & Kontoghiorghes, E. J. (2006), ‘Branch-and-bound algorithms for computing the best subset
regression models’, Journal of Computational and Graphical Statistics 15, 139–156.
Golub, G. H. & Van Loan, C. F. (1996), Matrix computations, Johns Hopkins Studies in the Mathematical Sciences, 3rd edn, Johns Hopkins University Press, Baltimore, Maryland.
Hofmann, M., Gatu, C. & Kontoghiorghes, E. J. (2008), ‘An exact least-trimmed-squares algorithm for
a range of coverage values’, Journal of Computational and Graphical Statistics . Submitted.
Hubert, M., Rousseeuw, P. J. & Aelst, S. V. (2008), ‘High-breakdown robust multivariate methods’,
Statistical science 23(1), 92–119.
Kontoghiorghes, E. J. (2004), ‘Computational methods for modifying seemingly unrelated regressions
models’, Journal of Computational and Applied Mathematics 162(1), 247–261.
Kontoghiorghes, E. J. & Clarke, M. R. B. (1995), ‘An alternative approach for the numerical solution
of seemingly unrelated regression equations models’, Computational Statistics & Data Analysis
19(4), 369–377.
Paige, C. C. (1978), ‘Numerically stable computations for general univariate linear models’, Communications in Statistics Part B — Simulation and Computation 7(5), 437–453.
Paige, C. C. (1979a), ‘Computer solution and perturbation analysis of generalized linear least squares
problems’, Mathematics of Computation 33(145), 171–183.
Paige, C. C. (1979b), ‘Fast numerically stable computations for generalized linear least squares problems’, SIAM Journal on Numerical Analysis 16(1), 165–171.
Rousseeuw, P. J., Aelst, S. V., Driessen, K. V. & Agulló, J. (2004), ‘Robust multivariate regression’,
Technometrics 46, 293–305.
Rousseeuw, P. J. & Leroy, A. M. (1987), Robust regression and outlier detection, John Wiley & Sons.
Rousseeuw, P. J. & Van Driessen, K. (2006), ‘Computing LTS regression for large data sets’, Data
Mining and Knowledge Discovery 12, 29–45.
Srivastava, V. K. & Dwivedi, T. D. (1979), ‘Estimation of seemingly unrelated regression equations
models: a brief survey’, Journal of Econometrics 10, 15–32.
16
Srivastava, V. K. & Giles, D. E. A. (1987), Seemingly unrelated regression equations models: estimation and inference ()p, Vol. 80 of Statistics: textbooks and monographs, Marcel Dekker, Inc.
Yanev, P. & Kontoghiorghes, E. J. (2007), ‘Computationally efficient methods for estimating the
updated-observations SUR models’, Applied Numerical Mathematics 57(11-12), 1245–1258.
Zellner, A. (1962), ‘An efficient method of estimating seemingly unrelated regression equations and
tests for aggregation bias’, Journal of the American Statistical Association 57, 348–368.
17