CROP 590 Experimental Design in Agriculture
advertisement

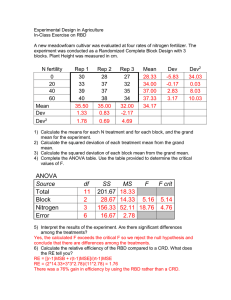
CROP 590 Experimental Design in Agriculture Lab exercise – 3rd week Two-Way Analysis of Variance RBD, missing values Power calculations SAS On-line Documentation SAS/STAT PROC GLM SAS/STAT PROC MIXED SAS/STAT PROC GLMPOWER We have seen how a one-way analysis of variance considers one treatment factor with two or more treatment levels. This type of analysis would be used for a Completely Randomized Design (CRD) experiment. Today we will go to the next level and look at the SAS commands for a Two-Way Analysis of Variance. We will analyze data from a Randomized Complete Block Design (RBD). We will also learn how SAS can handle missing data and estimate power for RBD experiments. A horticulturist performed a field experiment to determine the effect of fungicide treatments applied to plots where azaleas were to be grown. The treatments consisted of a control (no fungicide), an old product and a new product. The fungicides were applied to plots before inoculation in a Randomized Complete Block Design with 4 replications. Two plants were planted in each plot after inoculation. After several weeks the plants were dug up and their root weights were recorded. 1. Download the data for this exercise from the file Lab3.xlsx. Part I. RBD using plot means 2. For this analysis, we will work with the means for each plot (a conventional RBD design). The data are completely balanced (there are no missing plots). Write a SAS program to input data from the spreadsheet called “Plot Means” (in the Lab3.xls file). Use ‘input’ and ‘datalines’ statements and cut and paste the values from the spreadsheet directly into your SAS program. Remember that you need to include a $ sign after a character variable in your input statement, and that you need a semicolon at the end of your data lines. 3. Now use PROC GLM to run an RBD analysis on this data set. You will now have two independent variables in your model statement. The data are balanced (all treatments occur once in each block), so we can use the MEANS statement to obtain treatment means and request an LSD test. Proc GLM; Title 'RBD for fungicide treatments'; class block fungicide; model Rootwt=block fungicide; means fungicide/lsd; Run; 4. Interpret the results of the SAS output. Are there differences among the fungicide treatments? How does the new fungicide compare with the old product and the control? Was it advantageous to use blocks? 1 Part II. RBD with missing values 5. Now let’s compare the output when one or more of the means are missing. Make a copy of your SAS program, but replace the value of RootWt for the Old fungicide in Block 1 with a period (.). That is the designation for a missing plot in a SAS program or data text file. When you use the import wizard to bring in an excel spreadsheet, the missing cells should be left empty. SAS will automatically convert the missing cell to a ‘.’. 6. PROC GLM has the capability to make adjustments for missing values. However, you will need to replace the ‘Means’ statement with ‘LSMEANS’ (least squares means). It will no longer be possible to run a single LSD test for all treatment comparisons, because the standard errors will be slightly different for each mean and mean comparison. Instead, we will request standard errors (stderr) and the t statistics and probabilities associated with individual mean comparison tests (tdiff). If you only need the probability values, use the pdiff option rather than tdiff. Proc GLM; Title 'RBD with missing plots'; class block fungicide; model Rootwt=block fungicide; lsmeans fungicide/stderr tdiff; 7. A predicted value is the value of the response variable that is expected for a given treatment level based on the model. A residual is the difference between the observed and predicted value. To obtain estimates of predicted values and residuals (which can be plotted against the predicted values to check normality assumptions), include the following statements at the end of PROC GLM: Output out=new P=Yhat R=Resid; Proc Print data=new; run; Although we really have no reason to estimate a value for a missing plot in the modern era, you may be interested to know that the use of a missing plot formula would give you the predicted value in the case where you have a single missing observation in an RBD. Predicted values and residuals can now be found in the data set “new” in the “work” directory. Alternatively, you could create an output data set using the ODS system: ods output LSMeans=adjmean Diff=ptable PredictedValues=Yhat; This will create three new data sets: “adjmean”, “ptable” and “Yhat”. Open these data sets and note the contents of each. For a complete list of possible ODS table names, go to the GLM help documention, select the “details” tab, and choose “ODS Table Names”. 2 8. According to this analysis, are there significant differences among the fungicide treatments? Is the new fungicide different from the old? Were the block effects significant? Observe what happens to the Sums of Squares, df, the standard errors of means and the mean comparison tests when you have missing plots. Also note that with unbalanced data there is a difference between Type I and Type III Sums of Squares. In general, you will use the Type III Sums of Squares when you have missing data. 9. For unbalanced data, you should consider using a Mixed Model analysis if your model consists of some random (e.g. blocks) and some fixed effects (e.g. fungicides in this experiment). The syntax is similar, except that the random effects are not included in the model statement. Proc Mixed data=two; Title 'RBD with mixed models'; class block fungicide; model Rootwt=fungicide; Random block; lsmeans fungicide/tdiff; run; For balanced data, the results of the F tests of fixed effects for a Mixed Model analysis and ANOVA will be the same. Pairwise comparisons of treatment means will also be the same for the Mixed Model analysis and the ANOVA when data are balanced. Standard errors of means will be somewhat different for a Mixed Model analysis and an ANOVA, for both balanced and unbalanced data sets. Standard error estimates from Mixed Models will often be a little larger than from an ANOVA, because Mixed Models account for the additional variation due to sampling random factors (such as blocks) from a reference population. Part III. Power Calculations in an RBD 10. Use the estimate of MSE obtained from the ANOVA to conduct a power analysis: proc glmpower data=one; Title 'Power for fungicide experiment'; class Block Fungicide; model RootWt = Block Fungicide; power stddev = 1.518474 ntotal = 12 power = .; run; 3 Part IV. RBD using Excel 11. If time permits, try running a two-way ANOVA on the original data set (without missing plots) using Excel. First you must create a two-way table, with Blocks on one axis and treatments on the other. To run the analysis go to “Data” and select “Data Analysis” from the toolbar. (if your computer does not show the “Data Analysis” option you may have to add in the Analysis ToolPak - refer to your notes from Lab 2). Select “ANOVA: Two-factor without replication”. Excel considers blocks to be a second factor, and therefore does not recognize that the experiment is replicated. After you run the analysis, compare the results to SAS output from Part I. 4