Population Genetics The study of naturally occurring genetic differences among organisms
advertisement

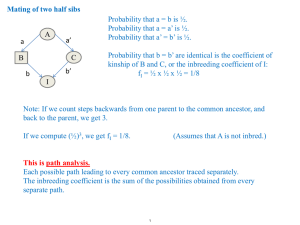
Consortium for Comparative Genomics University of Colorado School of Medicine Population Genetics The study of naturally occurring genetic differences among organisms Biochemistry and Molecular Genetics Consortium for Comparative Genomics Human Medical Genetics, Computational Bioscience University of Colorado School of Medicine David.Pollock@UCDenver.edu www.EvolutionaryGenomics.com Sexual reproduction What is Sex? Mixing Gender Ploidy Recombination Multiple Loci: Linkage 2 loci, A and B, in a good population, each in HW equilibrium The gametes may have nonrandom associations Multiple Loci: Linkage Gametes are A1B1, A1B2, A2B1, and A2B2 A1 B1 A1 B2 P11 P12 A2 B1 P21 A2 B2 P22 Allele frequencies of A and B, separately, are p1 and p2 for A, q1 and q2 for B Linkage and Recombination A new mutation is in disequilibrium with existing variants at other loci It arose linked to one variant or the other Linkage is disrupted through genetic exchange (breakage and reunion of gametes) Recombination, frequency r proportion of gametes with combination of alleles not present in either parent Range is 0 to 0.5 Linkage Disequilibrium Measurement of the degree of nonrandom association D = P11 P22 – P12 P21 Linkage and Recombination r determines the rate of approach to linkage equilibrium P11’ = r p1 q1 + (1-r) P11 = (1-r) D In each gen, D is only (1-r) times as large Falloff in LD with Distance Linkage and Time Starting with initial D0 Dt = D0 (1-r)t Values of D depend on allele frequencies Difficult to compare from one locus to the next Sometimes expressed as percentage of its minimum (if negative) or maximum (if positive) value E.g., Dmin = larger of –p1q1 and –p2q2 Incest is Best Riff Raff Mating between relatives Increased frequency of homozygotes Consider selfing (plants) AA x AA => AA aa x aa => aa Aa x Aa => 0.5 Aa + 0.25 AA + 0.25 aa Magenta Incest is Best Riff Raff Magenta Inbreeding coefficient AA: p2(1-F) + pF Aa: 2pq(1-F) aa: q2(1-F) + qF H0 H F H0 Incest is Best Identical by Descent (IBD) Relative to some ancestral population F is the probability that alleles are IBD Autozygous (always homozygous) Allozygous Not IBD; may be heterozygous or homozygous The difference can be important! Example of T1D and MHC alleles Inbreeding Depression Well, maybe not always best If individuals in normally outcrossed species are inbred, inbreeding harmful Data from Neal, 1935 Average Yield of Corn (Bushels/acre) 70 20 0 0.25 0.5 0.75 1.0 Inbreeding Coefficient, F Inbreeding Depression Well, maybe not always best Frequency of homozygous recessives for rare deleterious allele in first cousin mating F = 1/16 aa: q2(1-F) + qF aa: q2(1-1/16) + q/16 Relative Risk (RR) the The rarer the allele, greater the RR 1 1 q 1 q 16 16 RR 2 q 2 0.0625 RR 0.9375 q q 0.01, RR 700% Regular Systems of Mating Selfing, Sib mating, half-sib mating, or backcrossing to produce strains Calculate F at any time Hybrid vigor (heterosis) F => 0 after one generation of outcrossing Inbred lines unlikely to have same inbred rare deleterious recessive variants And the worst get weeded out Uniform progeny test for the best hybrids Mutation & Fate Absorbing States Extinction Fixation Ideal Population Constant N Hardy-Weinberg Fitness Fitness landscape Two pictures of evolution Adaptionists: (Dawkins, etc.) Neutralists (Kimura, Gould) Every day, in every way, I'm getting better and better! - Emile Coue “Nearlyneutral model” The Panglossian Paradigm The Spandrels of San Marcos A Force to be Reckoned With “Mankind is so unstable a species that he may one day become obsolete.” “This game is an exercise In controlling genetic drift.” Remote Inbreeding Getting ready for the coalescent IBD of any two alleles in a finite population Often FST to distinguish it Go back 1 generation, N alleles, a1…aN Probability that two alleles are the same 1/2N => the force of genetic drift Probability that two alleles are different, but related due to previous remote inbreeding (1-1/2N)Ft-1 1 1 Ft 1 F0 1 2N t Heterozygosity Requires Mutation Mutation breaks the chain of IBD Mutation rate m Probability no mutation = 1-m Infinite Alleles Model (all mutants different) 1 1 2 Ft 1 Ft11 m 2N 2N At Equilibrium Ft Ft1 Fˆ Fˆ 1 4Nm 1 4N m Hˆ 1 Fˆ 4Nm 1 1 Effective Population Size The population size of an ideal population that would produce the same outcome (i.e., same heterozygosity, given mutation rate) 4Ne m Variance in expected reproduction, changing population size, male to female ratios (and different mating success), dispersion (neighborhood size) all reduce the effective population size Migration Migration limits genetic divergence Remarkably little: 1-4 migrants per generation (Nm) Low migration rates is like increased remote inbreeding Apparent inbreeding FST Due to population substructure Variance over expected variance Happens even though each subpopulation is undergoing random mating, in HWE 2 pq The Driving Force Natural Selection Inherited differences in the ability of organisms to survive and reproduce Through time, better genotypes increase in frequency in a population Natural selection chooses between different variants Variants compete with one another Leads to greater adaptation of organisms to their environment Accumulation of ability to survive and reproduce Strategies to Produce More Offspring Survival (zygotic selection) Fertility (mating success, sexual selection) Fecundity (production of zygotes) Gametic selection (distorted segregation in zygotes) Parenting (increase the probability that your young will survive) There are usually trade-offs E.g., More offspring at younger age = lower survival Mostly Competition Between Individuals Modeling Natural Selection AA p AA aa q aa 2 2 Aa 2pq Aa 2 p AA 2 pq Aa q aa 2 f xy f xy xy Dominant Recessive From CA Andrews, 2010 Selection Needs to Overcome Genetic Drift 10 replicates, N=20 From CA Andrews, 2010 Predictions Mutation-selection balance Time required for changes in gene frequency Overdominance, underdominance, frequencydependent selection (e.g., MHC) Selection versus drift Diffusion approximations Effective population size The genetic architecture of complex traits Phenotypic variation due to population average plus Genotypic variance Environmental variance Interaction between the two Heritability: ratio of genotypic to phenotypic variance Depends on variant frequencies! Quantitative traits, multiple genes Pleiotropy (antagonistic?) During a drought, Finch N decreased due to decline in seed supply. Size and hardness of seeds increased, as did the size of the finches (larger birds can eat bigger seeds)