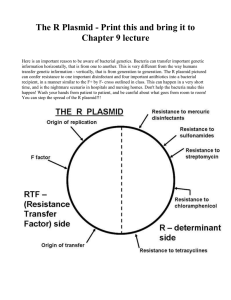
Section I: Purpose and Scope of Effort
advertisement

Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Contract No. HHSN266200500040-C ADB Contract No. N01-AI-50040 Section I: Purpose and Scope of Effort The Tularemia Vaccine Development Contract will lead to vaccine candidates, two animal models and cellular assays vital for testing vaccine efficacy. Sections II and III: Progress and Planning Presented by Milestone Active milestones: 2, 3, Working Group, 5, 12 (UNM &LBERI), 25, 26, 32, 33, 39, 40, 41, 43, 46, 49, 50 Completed milestones: 1, 16 Inactive milestones: 4, 6-11, 13-24, 27-31, 34-38, 42, 44-45, 47-48, 51-54 Milestone 2 Milestone description: Vaccinations performed on relevant personnel Institution: UNM/LRRI 1. Date started: 11/01/1005 2. Date completed: pending 3. Work performed and progress including data and preliminary conclusions a. NIAID is working on the IAA with USAMRIID and a legal and financial liability review is pending. b. Dr. Bob Rubin, President/CEO of LRRI, is discussing access to the LVS vaccinations with Dr. Ed Nuzum ,who is the Chief of the Product Development Section in the Office of Biodefense Research Affairs at NIAID. Dr. Rubin wants the LVS vaccine for the LRRI scientists working on tularemia. LRRI wants best protection for their team and is willing to waive the indemnification issue for their scientists. c. Another option may be through LRRI’s clinical trials subsidiary that performs I&D for different funding agencies. Could LRRI be a site for giving the LVS vaccine through LRRI’s subsidiary? Dr. Chuck Hobbs, of LRRI, was asked by Dr. Ed Nuzum to perform a cost analysis. 4. Significant decisions made or pending a. UNM and NIAID continue to wait for a change in the status of the IAA between NIAID and USAMRIID. b. UNM and LBERI will use their biobubbles as additional physical protective equipment c. NIAID will need to provide UNM access to human cells from other LVS vaccinated individuals which are needed to develop in vitro immunoassays. For possibly another year, UNM will not have access to a local source of human cells from LVS vaccinated individuals d. UNM EOHS has obtained many of the laboratory documents i. Documents pending 1. Radiology Facility Accreditation Certificate 5. Problems or concerns and strategies to address UNM will need an external source of human cells from LVS vaccinated individuals, in order to develop the immunoassays in humans. 6. Deliverables completed None 1 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 7. Quality of performance Good 8. Percentage completed 16%- no change relative to 8/15/06 report 9. Work plan for upcoming month Ross Kelley will continue to monitor the progress of whether Martin Crumrine's IAA between NIAID and USAMRIID will inform UNM when and whether the TVD Contractors can be vaccinated under this IAA. 10. Anticipated travel Travel could occur in September 2006 to September 2007, depending on the completion of the IAA. 11. Upcoming Contract Authorization (COA) for subcontractors UNM may request a COA to allow 1-2 UNM EOHS nurses to travel to USAMRIID for training on LVS site vaccination evaluations. The timing of the COA request depends on the achievement of the IAA. Milestone 3 Milestone description: Bioaerosol technique selected and optimized Institution: LBERI 1. Date started: 2/23/2006 2. Date completed: in progress 3. Work performed and progress including data and preliminary conclusions Created a new stock of frozen LVS from Chamberlain’s (8/11/06) and titered the stock at 1.6x108 cfu/mL on CHAB. This stock is being used to perform aerosol sprays with thawed LVS. Performed 3 additional bioaerosol sprays with the Collison nebulizer and thawed LVS; one experiment (8/8/06) compared the relative efficiencies of the AGI (all glass impinger) and the biosampler for quantitating viable recovery of LVS; one experiment (8/15/06) was set up to determine the lower limit of LVS aerosol exposure; one experiment (8/25/06) determined the reproducibility of the lower limit of LVS aerosol exposure. Data are shown below. Experiment 8/8/06 performed with thawed LVS demonstrated that the AGI efficiency was best. Experiment 8/15/06 suggested that the low limit for LVS aerosols is approximately 1x10 4 CFU/mL in the spray. Below 1x104 CFU/mL in the spray resulted in less than 5-10 colonies per plate, which is too low due to potential variability. Experiment 8/25/06 replicated the data from 8/15/06 and demonstrated that the lower limit for LVS aerosols is approximately 1x104 CFU/mL in the spray Data files are found in \\Saturn\ABSL3\Study Data (2005-2006)\Francisella tularensis\FY06078 (Tul-03)\Bioaerosol Data Experiment 8/8/06. Comparison of AGI and Biosampler Efficiency Sampler AGI Biosampler Approximate CFU/mL sprayed (N=3) 2x108 CFU/mL Recovered (N=3) 1.26x105 CFU/L (N=3) 4.66x104 Spray Factor (N=3) 3.94x10-7 2x108 1.33 x105 2.16 x104 1.67 x10-7 2 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Experiment 8/15/06. Determination of Lower Limit for LVS Bioaerosols Target Spray Conc (CFU/mL) 5x103 1x104 5x104 1x105 5x105 Actual Spray Conc. (CFU/mL) 5.67E+03 1.29E+04 8.83E+04 1.68E+05 6.80E+05 CFU/mL Recovered 1.00E+01 5.33E+01 2.77E+02 1.40E+02 1.00E+03 CFU/L Spray Factor 3.71E+00 1.65E+01 1.00E+02 5.13E+01 3.71E+02 6.54E-07 1.28E-06 1.14E-06 3.06E-07 5.45E-07 Experiment 8/25/06. Determination of Lower Limit for LVS Bioaerosols (Replicate Sprays) Target Spray Conc (CFU/mL) 1x104 3x104 5x104 No. of Replicates 3 3 3 Actual Spray Conc. (CFU/mL) 2.52E+03 9.43E+03 1.87E+04 CFU/mL Recovered 1.89E+01 2.33E+01 5.56E+01 CFU/L 6.98E+00 8.59E+00 2.02E+01 Spray Factor 5.42E-06 9.66E-07 1.08E-06 4. Significant decisions made or pending a. None 5. Problems or concerns and strategies to address a. None 6. Deliverables completed a. None 7. Quality of performance a. Good 8. Percentage completed a. 22% 9. Work plan for upcoming month Perform bioaerosol experiments on vegetative LVS with Collison generator to compare with thawed LVS Repeat of studies performed on thawed LVS, but now with vegetative. The studies will include measurement of spray factors, reproducibility, high and low doses. Plan to grow LVS in CB Plan to quantitate LVS on CHAB 10. Anticipated travel a. None anticipated at the present time 11. Upcoming Contract Authorization (COA) for subcontractors a. None anticipated Working Group Milestone description: Determine appropriate solid and liquid media for growth of tularemia for project team Institution: LBERI/UNM 1. Date started: 2/23/2006 2. Date completed: in progress 3. Work performed and progress including data and preliminary conclusions: 3 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble a. None for LBERI; UNM is performing virulence studies with DVC Lot#16 LVS grown in Chamberlains versus from lyophilized vial. See UNM MS 5 below. 4. Significant decisions made or pending a. A meeting of the working group examined blue/grey colony morphology data produced by DSTL and determined that Chamberlain’s broth and CHAB agar outperformed the other media tested and recommended that these media be used in growth of LVS stocks. 5. Problems or concerns and strategies to address a. None 6. Deliverables completed a. Determined liquid and solid media for LVS growth 7. Quality of performance a. Good 8. Percentage completed a. 100% for LBERI; UNM needs to report a percentage for the virulence studies. 9. Work plan for upcoming month a. UNM is performing experiments to compare the virulence of LVS grown in Chamberlain’s media with the virulence of freshly reconstituted LVS from a vial of DVC lot#16 LVS. 10. Anticipated travel a. None anticipated at the present time 11. Upcoming Contract Authorization (COA) for subcontractors a. None anticipated Milestone 5 Milestone description: Species tested for sensitivity to LVS & generation of immunity against a pulmonary challenge of Schu4 Institution: UNM 1. Date started: 12/12/2005 2. Date completed: pending 3. Work performed and progress including data and preliminary conclusions a. Experiment Ftc12, study 8 (notebook 85, pages 20-23), study 9 (notebook 85, page 29), study 10 (notebook 85, pages 39-41) i. The purpose was to develop a method to infect Fischer 344 rats by the pulmonary route. ii. The major obstacle up to now has been the difficulty visualizing the trachea. This may be attributed in part to the ketamine/xylazine anesthesia that was used [CF Schaefer et al. J Appl Physiol. (1984) 56:533-5]. Since isoflurane does not have these effects, we decided to use it instead. iii. Our intention has been to infect rats using the microspray aerosolizer from Penn Century. However, there are several disadvantages to using this system 1. In a side-by-side in vitro comparison, the amount of bacteria delivered by the microspray aerosolizer was only 25% of that delivered by a 200 l pipetmen. It is possible that the microspray aerosolizer delivered a smaller volume or that it generated enough shear forces to kill F. tularensis. 2. The microspray aerosolizer can only deliver five 50 l or two 100 l doses. It will therefore be necessary to frequently reload the aerosolizer for large experiments. However, the technique for 4 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble reloading the aerosolizer makes it very likely to contaminate all work surfaces. iv. We are evaluating an alternative method of intratracheal inoculation using a blunted-ended, 18-gauge, 1¾” i.v. catheter. 1. Rats were anesthetized with isoflurane 2. The plastic sheath from the i.v. catheter was inserted into the trachea and proper placement was verified by pumping a small volume of air into the lungs; if the sheath had been inserted into esophagus, there would be a vacuum when drawing air back into the syringe. 100 l of the inoculum was then delivered with the blunt-ended catheter needle attached to a disposable 1 ml syringe and followed by 200 l of air to ensure the entire inoculum was delivered. 3. We successfully inoculated 100% (n=4) Fischer 344 rats using this method a. The target dose was 6.12 x 105 CFU LVS/rat b. An average of 3.1 ± 0.5 x 105 CFU/rat (49% of initial inoculum) was recovered from lung homogenates. 4. We will train additional members of the team on this method b. Experiment Ftc17 (Notebook 85, pages 44-47) i. The purpose was to determine the intranasal LD50 of SCHU S4 in s.c.-, i.d.-, and i.n.-LVS vaccinated NIH-Swiss and BALB/c mice. ii. Two months after vaccination, 2 mice from each group were killed and shown to have completely cleared LVS from the lungs, spleens, and livers iii. I.n.-vaccinated mice were challenged i.n. with 51, 230, and 1640 CFU SCHU S4 iv. I.d.- and s.c.-vaccinated mice were challenged i.n. with <20, 20, and 40 CFU SCHU S4. v. We are monitoring clinical symptoms and survival c. Experiment Ftc19 study 1 (Notebook 85, pages 30-31), study 2 (Notebook 85, pages 32-33), study 3 (Notebook 85, pages 34-35) and study 4 (42-43) i. The purpose was to generate a LVS working stock from DVC’s lot 16 in Chamberlain’s medium using the exact conditions established by the TVDC working group. This stock will then be used for all future TVDC experiments ii. The culture conditions were: 1. Rehydrate LVS in 1 ml sterile PBS 2. Inoculate 100 l into 100 ml Chamberlain’s medium in a 250 ml disposable, screw-top, Erlenmeyer flask 3. Shake culture for 48 h at 37oC and 225 rpm iii. In Ftc19 study 1, it took four days for OD600 to reach 0.391. This was unacceptable because the TVDC working group indicated that after 2 days in culture a blue-gray phase shift is likely to occur. iv. In Ftc19 study 2 and study 3, no growth was observed v. We suspected that the Chamberlain’s powder may be light sensitive since its activity decreased over time and it was stored in a clear tube. Therefore, we obtained a second aliquot of Chamberlain’s powder from Bob Sherwood. This aliquot had been stored in the dark by wrapping in foil. vi. In Ftc19 study 4, the 48 hour OD600 was 0.644, equivalent to 2-3 x 109 CFU/ml vii. 291 aliquots (300 l each) were created and stored without preservatives in a –80oC freezer 5 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble viii. In Ftc19 study 5, we will compare the virulence of LVS grown in Chamberlain’s medium to that of DVC’s lot 16 LVS and will share the data with the Working Group 4. Significant decisions made or pending For the rat model, we will use isoflurane rather than ketamine/xylazine anesthesia and use an iv catheter rather than a microsprayer for the intratracheal delivery of inoculations. Powdered Chamberlain medium will be stored in the dark and kept dry. 5. Problems or concerns and strategies to address None 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed 14% 9. Work plan for upcoming month a. Determine the intranasal LD50 of SCHU S4 in s.c.-, i.d.-, and i.n. vaccinated NIHSwiss and BALB/c mice b. Compare the virulence of LVS grown in Chamberlain’s medium to DVC’s lot 16 LVS and report the results to the Working Group. c. Determine the sensitivity of Fischer 344 rats to pulmonary SCHU S4 infection d. Determine the resistance of s.c.- and i.d.-vaccinated Fischer 344 rats to a lethal pulmonary SCHU S4 infection e. Start training on handling, anesthetizing, and infecting guinea pigs 10. Anticipated travel a. 9/26-9/27/06: Annual meeting in Albuquerque, NM b. 10/30 to 11/4/06: Rick Lyons, Terry Wu and Barbara Griffith to attend Collaborative meeting with DVC and NIAID and to attend 5th International Tularemia Conference in Woods Hole MA 11. Upcoming Contract Authorization (COA) for subcontractors COA #11: authorizes travel for 7 UNM TVDC scientists to the 5th International Tularemia Conference in Woods Hole, MA Milestone 12-UNM Milestone description: Assays for detecting relevant immune responses in animals & humans developed Institution: UNM 1. Date started: 7/15/06 2. Date completed: Pending 3. Work performed and progress including data and preliminary conclusions a. Ftc18 study 2 (Notebook 85, pages 25-28) b. The purpose was to determine the best antigen for measuring antigen-specific T cell responses c. Three method of antigen preparation (figure 1) i. Heat-killed - LVS and SCHU S4 were effectively killed by 3 h incubation at 70oC ii. Formalin-fixed – LVS and SCHU S4 were effectively killed by 30 min incubation in 3% or 10% neutral-buffered formalin with or without 60oC iii. UV-inactivation 6 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 1. SCHU S4 was effectively killed by 60 min incubation 2. One LVS colony was detected after 60 min incubation. A longer incubation period (90 min) may be required 4. Significant decisions made or pending NA 5. Problems or concerns and strategies to address We had initially planned to use the loss of light-emitted by LVS-luciferase as a convenient readout for killing of intracellular bacteria. However, the assay sensitivity is low and the dynamic range is limited. Therefore, we will measure killing of intracellular bacteria by plating cell lysates onto cystine heart agar and incorporate more convenient readout when they become available in the future. 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed 3% 9. Work plan for upcoming month 7 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble a. Determine whether splenocytes (including macrophages and T cells) from LVS vaccinated BALB/c mice can kill F. tularensis in a co-culture system. The detection assay will be based on plating and quantification of CFU. b. Determine the best antigen (heat-killed, formaldehyde-fixed, or UV-inactivated F. tularensis) for stimulating proliferation and cytokine production by antigen-specific T cells from vaccinated mice 10. Anticipated travel a. 9/20-9/21/06: Alexandra Scrymegeour and Tara Hendry-Hofer to Fisher Immunoassay training course in Rockville, MD 11. Upcoming Contract Authorization (COA) for subcontractors COA# 10: authorizes travel to the CMI Workshop in Rockville MD. Milestone 12-LBERI Milestone description: Assays for detecting relevant immune responses in animals & humans developed Institution: LBERI 1. Date started: 2/23/2006 2. Date completed: in progress 3. Work performed and progress including data and preliminary conclusions a. Purpose: Repeat protocol from the Purdue Cytometry ListSERV using Nycomed Lymphoprep ,which to date provided the highest yield of peripheral blood mononuclear cells (PBMCs) from cynomolgous macaque blood, not contaminated by RBCs, i. Goal was to determine whether this protocol gave a high yield of cells which were not highly contaminated by RBCs, and that responded vigorously and specifically to immune stimulation (mitogenic initially and later, antigenic) ii. Raw data: TVDC Binder 1 (Wilder Lab), TUL-3 (8/14/06; repeat of Purdue Cytometry Nycomed Lymphoprep method for purification of cynomolgous macaque PBMCs); C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\tul3 elispot 081606.xls C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\Tul 3 prolif 081806.xls iii. Summary data: C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\TUL3 summary.xls C:\Documents Settings\jwilder.LOBOS\My Documents\Tularemia Contract\tul3 elispot 081606 incl summary.xls C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\PBMC assays.svv b. Methods for purification i. Brief outline of the Purdue Cytometry Lymphoprep protocol 1. Dilute blood 1:3 with PBS 2. Layer 2 parts blood on top of 1 part Lymphoprep 3. Spin 30 minutes at RT, no brake, 1800 RPM 4. Harvest interface and wash twice with PBS 5. Lyse RBCs, spin and count pellet ii. Full protocol can be found in TVDC Binder 1 (Wilder Lab) c. Results for yield and purity 8 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble i. Summary of results shown below for TUL3 in comparison to Tul 1 and 2 (C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\tul 1-3 summary tables 090606) ii. Tables show that the Purdue CML protocol proved superior in resultant yield (20-95 x 106/~4.3ml, and RBC contamination (7-10%) both times that we have tested it d. Methods for proliferation assay with mitogenic stimuli i. Cells were set up in a proliferation assay to test their response to mitogenic stimuli 1. Tested their response to Con A (10 g/ml) and anti-CD3 (from the MABTECH ELISpot kit (#3420M-2HW-Plus); 100 ng/ml) 2. Cells were cultured in quadruplicate for 4 days before addition of BRDU; BRDU was left in for 18 hours before development of the anti-BRDU ELISA as per kit instructions (Cell Proliferation ELISA, BrdU (chemiluminescent), Roche Applied Science, Cat. # 11 669 915 001) e. Results for proliferation assay with mitogenic stimuli 1. observed good proliferative capacity to Con A, but not anti-CD3, at three different cell concentrations (0.5 x 106/ml, 1 x 106/ml and 1.5 x 106/ml); see below TUL3 summary.xls “Cyno PBMC Proliferation: Tul3” 2. . Proliferation data shown below and also in TVDC Binder 1, Tul 3 a. Raw data: C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\ Tul 3 prolif 081806.xls b. Summary data: C:\Documents and Settings\ jwilder.LOBOS\My Documents\Tularemia Contract\ TUL3 summary.xls f. Methods for IFN response to mitogenic stimuli i. Cells were set up in a IFN ELISPOT assay to test their response to mitogenic stimuli 1. Tested their response to Con A (10 g/ml) and anti-CD3 (from the MABTECH ELISpot kit (#3420M-2HW-Plus); 100 ng/ml) 2. Cells were cultured initially overnight in polypropylene tubes, 1.5 x 106/ml at 37 degrees (while the ELISPOT plates were coated with antiIFN antibody – normally this would be done the night before and the cells would be cultured directly on these plates) 3. The following day, the ELISPOT plates were washed and blocked with culture medium as per the kit instructions 4. Cells were transferred from tubes to the plates, in quadruplicate, at the various densities/well: 225,000, 112,500, 56,250, 28,125 and 14,063 5. Plates were incubated a further 20.5 hours, a total of 38.5 hours with the mitogens 6. Plates were washed, anti-IFN-biotin detection antibody was added at 1 g/ml for 2 hours 7. Plates were washed and strept-avidin-HRP added for 1 hour 8. Plates were washed and TMB substrate added and incubated 20 minutes before washing extensively under tap water 9. Plates were left to air dry and read on the ELISPOT reader at UNM on 8/18/06 g. Results for IFN response to mitogenic stimuli i. Results are shown below, TUL-3: ELISPOT results: 1. saw good stimulation of IFN production after Con A stimulation, but not anti-CD3 2. Con A stimulation of wells containing more than 28,125 cells resulted in spots that were too dense to enumerate 3. Data locations: 9 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble i. Raw data: C:\Documents Settings\jwilder.LOBOS\MyDocuments\Tularemia Contract\ tul3 elispot 081606.xls C:\Documents and Settings\jwilder.LOBOS\MyDocuments\Tularemia Contract\PBMC assays.svd ii.Summary data: C:\Documents and Settings\ jwilder.LOBOS\My Documents\Tularemia Contract\tul3 elispot 081606 incl summary.xls C:\Documents and Settings\jwilder.LOBOS\My Documents\Tularemia Contract\PBMC assays.svv Tul 1-3 summary tables 090606.doc Yield and Purity of PBMCs Date 7/12/06 Protocol Purdue CML Animal F2519 7/24/06 Williams’ Lab 2146C 7/24/06 NHPRR 55DIE 8/14/06 Purdue CML 63909 Yield 19.4 x 106/4.2 ml blood 10.4 x 106/4.7 ml blood 0/4.8 ml blood 94.75 x 106/4.5 ml blood % Viable 74% % RBCs 10.7% > 80% Approx. 70.0% n/a n/a 82.8% 7.6% TUL3 Summary.xls Cyno PBMC Proliferation: Tul3 900000 800000 700000 RLU 600000 0.5 x 106/ml 1 x 106/ml 1.5 x 106/ml 500000 400000 300000 200000 100000 0 2255 15599 Nothing Media only Cells only Cells + ConA Cells + anti-CD3 Key to Graph: -Nothing: No cells in wells; no addition of BRDU or anti-BRDU or substrate to those wells -Media only: Media in wells for 4 days; addition of BRDU, anti-BRDU and substrate -Cells only: Unstimulated cells in wells; addition of all reagents to detect proliferation 10 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble TUL-3: ELISPOT Results Cell Mean for IFNg Spots 300 Media Con A Anti-CD3 250 200 150 100 50 0 14063 28125 56250 112500 225000 Number of Cells Plated/Well d. Summary/Conclusions i. Purdue cytometry mailing list protocol is superior for yield and low RBC contamination 1. Cells respond well to Con A as measured by proliferation and IFN secretion iv. IFN ELISPOT kit is simple and sensitive, however, the positive control provided, anti-CD3, does not stimulate the cyno PBMCs well at 100 ng/ml 4. Significant decisions made or pending Made: Use of Purdue Cytometry Mailing List (PCML) protocol employing Nycomed Lymphoprep separation media 5. Problems or concerns and strategies to address None 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed 60% of scientific work has been completed 9. Work plan for upcoming month 1) We will prepare two batches of cynomologous macaque PBMCs from the same animal using the PCML protocol and stimulate them in culture with Con A and PHA to test proliferation and IFN secretion; this will test how reproducible the procedure is from isolation to isolation. 2) We will stain the whole blood and PBMCs with antibodies to detect T cells (anti-CD4 and –CD8), monocytes (anti-CD14) and NK cells (anti-CD56) to begin to assess the distribution of these cells in normal cyno blood . 3) We will obtain expert help from the cytologists at LRRI who are very familiar with cynomolgous macaque differential analysis to ascertain whether we are accurately reading 11 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble our slides and calling the correct percentage of lymphocytes, macrophages, eosinophils and neutrophils in our cyno PBMC preparations. 10. Anticipated travel None anticipated at the present time 11. Upcoming Contract Authorization (COA) for subcontractors None for this milestone. Milestone 25 Milestone description: Design protein-fragment library based on SCHU S4 sequence Institution: ASU-Sykes 1. Date started: 3/02/2006 2. Date completed: Pending 3. Work performed and progress including data and preliminary conclusions Pilot studies designed to evaluate our new synthetic-gene prediction program and new assembly protocols are nearly complete. These protocols uniquely enable us to take advantage of the newly developed microfluidic-based synthesis technologies for oligos. These new oligonucleotides are parallel-synthesized and therefore are significantly less expensive than standard individual-resin synthesized ones. However, oligo scale and quality is lower, and oligos are delivered as an oligomix instead of individually arrayed. We have had 2 breakthroughs in the past couple weeks. First, we have successfully assembled multiple subgene “building blocks” of the intended 250bp size into full genes with high sequence fidelity. In fact, the gene quality is greater than that from standard oligos. The second breakthrough is the successful design and production of larger”building blocks” of 500bp each. This should further reduce production costs since only half as many block amplifications will be necessary. The list of 500 synthetic peptides covering MHC Class I and Class II epitopes have been reviewed by Drs. Sykes, Johnston, Lyons, and Breen. The order has been placed but has not yet been received. Files are stored at R:\GeneVac\FTU\Contract\Proteome\Milestones\26\MHC\ftu_mhc_output.xls 4. Significant decisions made or pending Examine fidelity, biases, and yield of assembled genes generated with micro-chip vs. resin-based oligo-synthesis technologies. Fidelity will be assessed by cloning representative ORFs and individually sequencing them. ORF assembling and/or amplification biases will be assessed by comparing relative yields of sets of building blocks derived from a single oligomix. ORF yields will be determined by nanodrop spectrophotometry. 5. Problems or concerns and strategies to address None. 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed 85% 9. Work plan for upcoming month a. Select final oligo design for library based on ORF product quality and quantity. b. Receive, dilute, and array pools of peptides for sending to UNM for in vitro testing. 10. Anticipated travel Travel to UNM on September 26-29 for annual TVDC meeting and WRCE meeting. 11. Upcoming Contract Authorization (COA) for subcontractors None 12 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Milestone 26 Milestone description: Confirmation of gene expression (Design HTP SOPs, Test HTP SOPs, ORF library production, confirm gene expression) Institution: ASU-Sykes 1. Date started: 3/02/2006 2. Date completed: Pending 3. Work performed and progress including data and preliminary conclusions We have designed and received primers for amplifying 7 targeted F. tularensis genes (Table 1). The ORF products have been amplified (Figure 1.) Table 1 Selected F. tularensis genes for IVT production 1. FTT0208c ABC transporter 2. FTT1695 groES chaperone 3. FTT1712c 23 kDa protein 4. FTT0901 TUL4 5. FTT1419 hypothetical lipoprotein (p11) 6. FTT1602 hypothetical lipoprotein (p12) 7. FTT0613c hypothetical membrane protein (p15) Figure 1. PCR amplification of selected F. tularensis genes in Table 1 Files are stored at R:\GeneVac\FTU\Contract\Proteome\Milestones\26\ All 7 genes have been covalently placed in all four construct designs by overlapping PCR. 4. Significant decisions made or pending These 28 constructs are being evaluated for protein production based on a number of criteria. 13 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble These alternative constructs will allow us to select the optimal arrangement of ORF promoters, and protein fusions and tags for the library of antigens. 5. Problems or concerns and strategies to address None 6. Deliverables completed None. 7. Quality of performance Good 8. Percentage completed 65% 9. Work plan for upcoming month Comparatively evaluate IVT constructs in several reaction mixes. Select the optimal construct and IVT reactions based on ease of purification, yield and specificity of products and robustness of the protocol. 10. Anticipated travel Travel to UNM on September 26-29 for annual TVDC meeting and the WRCE meeting. 11. Upcoming Contract Authorization (COA) for subcontractors None Milestone 32 Milestone description: Oligos selected for microarray production; Oligos list refined, 70mer oligos procured, GDP oligos defined, Based on SCHU S4 sequence. Institution: ASU-Johnston 1. Date started: 3/02/2006 2. Date completed: 08/28/2006 3. Work performed and progress including data and preliminary conclusions The 70mer oligo set has been designed to cover the SCHU S4 genome; the set has been ordered and received. The GDP primers have been designed, ordered and received. 4. Significant decisions made or pending None 5. Problems or concerns and strategies to address None 6. Deliverables completed Oligonucleotide probes and controls (1,824, 70 mers) were arrayed into 5, 384 master and one working spotting plates for creating spotted microarrays to assess the gene expression profile of F. tularensis. Set of GDP primers for genome amplification. 7. Quality of performance Good 8. Percentage completed 100% 9. Work plan for upcoming month None 10. Anticipated travel Travel to UNM on September 26-27 for annual TVDC meeting. 11. Upcoming Contract Authorization (COA) for subcontractors None 14 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Milestone 33 Milestone description: Microarrays constructed and confirmed; First printing of arrays, Testing with DNA from Ft, Arrays GDPs validated at ASU Institution: ASU-Johnston 1. Date started: 08-01-2006 2. Date completed: Pending 3. Work performed and progress including data and preliminary conclusions We received 19, 96 well plates containing a minimum of 65 microliters of an 80 µM stock solution of each of the 1,824, 70mer oligonucleotides and controls. These have been re-arrayed into master and working 384 well plates for printing. A test plate of oligonucleotide probes has been prepared in either 3XSSC vs. Array-It Spotting buffer to determine optimal spotting conditions (Figure 2). The test plate has been used to print microarray slides which will be used for test hybridizations with LVS and/or SCHU S4 genomic DNA Figure 2. Red-Reflect image (salt deposition) of a subset of F. tularensis microarray probes in either 3XSSC or Array-It Spotting Buffer 4. Significant decisions made or pending Perform hybridization efficiency with genomic DNA in both spotting buffer conditions before final dilution of working plates to 40 µM in the selected spotting buffer. 5. Problems or concerns and strategies to address None 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed 30% 9. Work plan for upcoming month Perform QC analysis of received LVS and SCHU S4RNA and DNA samples from UNM. Perform hybridization tests using the two spotting buffer conditions with a limited set of probes. With this result, complete the dilution of the first working set and print test slides on both Corning Ultragaps 15 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble and in-house produced poly-L-lysine-coated slides. Begin amplification studies of diluted RNA with GDP primers for amplification efficiency. 10. Anticipated travel Travel to UNM on September 26-29 for annual TVDC meeting and WRCE meeting. 11. Upcoming Contract Authorization (COA) for subcontractors None Milestone 39 Milestone description: Create uvrA or uvrB mutant F. tularensis subsp. novicida Institution: UTSA 1. Date started:4/3/2006 2. Date completed: Pending 3. Work performed and progress The first part of the report will be a summary of the first deletion made in U112 (uvrB), reported earlier, followed by the new data which is the creation of uvrA deletion in U112 (KKF72). 3.1 The primers used for making UvrB mutants are: For UvrBUpSeq UvrBUp 5’ggaGAATTCctg tga gtg gtg tat ttg gct cga UvrBDn 5’ act act ggg ctg ctt cct aat gca ttg tat tgc ttg agg ctg atc gcc For UvrBDnSeq: UvBUp1 5’gct gct aac aaa gcc cga aag gaa gct acg aag gtt atc aaa gct ctc g UvrBDn1 5’ gga GAA TTC ttg cac caa tcc cgg caa gtaa For FnPErmCSeq pET15bUniUp: 5’tgcattaggaagcagcccagtagt pET15bUniDn: 5’ttc ctt tcg ggc ttt gtt agc agc 3.2 Set up following PCR reactions to amplify UvrBUpSeq: 10 X KOD XL Buffer 5.0 ul dNTP 2mM 5.0 ul UvrBUp 2uM 5.0 ul UvrBDn 2uM 5.0 ul U112 Chromosomal DNA 10 ng/ul 5.0 ul KOD XL DNA polymerase 0.4 ul dH2O 24.6 ul and UvrBDnSeq: 10 X KOD XL Buffer 5.0 ul dNTP 2mM 5.0 ul UvrBUp1 2uM 5.0 ul UvrBDn1 2uM 5.0 ul U112 Chromosomal DNA 10 ng/ul 5.0 ul KOD XL DNA polymerase 0.4 ul dH2O 24.6 ul at 94C 2’ then 94C 30”, 55C 30”, 72C 30” for 35 cycles, and at 72C for 10’. 16 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 3.3 The gel picture of UvrBUpSeq and UvrBDnSeq is shown as below: 6 5 4 3 2 1 Lane 1-3, UvrBUpSeq; Lane 4-6, UvrBDnSeq. 3.4 For overlapping PCR, set up: Set up: 10 X KOD XL Buffer 5.0 ul dNTP 2mM 5.0 ul UvrBUp 2uM 5.0 ul UvrBDn1 2uM 5.0 ul UvrBUpSeq 3.0 ul FpErmCSeq 3.0 ul UvrBDnSeq 3.0 ul KOD XL DNA polymerase 0.4 ul dH2O 24.6 ul at 94C 2’, then 94C 30”, 55C 30”, 72C 3’ for 35 cycles, and 72C 10’. 3.5 The gel picture was shown for overlapping PCR as below. 3 2 1 Lane 1-3, UvrBUpSeqFpErmCSeqUvrBDnSeq 3.6. Identify the UvrBFpEmrC inserted into pGEM-T vector. The fragment from Step 3.4 was inserted into pGEM-T. White colonies from Amp/X-gal plate was grown up, and plasmid was cut with EcoRI. 17 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble The gel picture was shown as below: 2 1 Lane 1,2 were plasmids cut with EcoRI with insert. 3.7 The plasmid from lane 1 was sequenced, and is correct. The plasmid was designated as pKEK 951. pKEK 951 is the uvrA:ermC deletion plasmid. 3.8 The pKEK 951 was cryotransformed into U112, and grown up on TSA/ErmC plate. Mutants were screened with colony PCR. The gel picture was shown as below. 9 8 7 6 5 4 3 2 1 Lane 1: Negative Control Lane2: U112 wild type control Lane 3: pKEK951 control Lane 4-9: Mutants. 3.9 The PCR fragments from Step 3.7 were purified and cut with BamHI. The gel picture was shown as below: 4 3 2 1 18 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Lane 1: Schu4 wild type template Lane 2: U112 wild type template Lane 3: pKEK951 template Lane 4: Mutant As only mutant has BamHI site, like plasmid pKEK951, which can be cut into two fragments. The PCR fragments of wild type can not be cut as there is no such site in both Schu4 and U112. 3.10 PCR fragment from step 3.8 was sequenced, and is correct. The mutant was frozen down and designated as KKF 71. KKF71 is the uvrB::ermC mutant in U112. 3.11 The primers used for making UvrA mutants are: For UvrAUpSeq UvrASchu4Up 5’GGAGAATTCTGAAGCTATAGCAGAGGCTCGTGA UvrASchu4LVSDn ’ACTACTGGGCTGCTTCCTAATGCACAACATACCTTCTTT GCCCTTCAG For UvrADnSeq : UvrASchu4LVSUp1 5’GCTGCTAACAAAGCCCGAAAGGAAGAAGGTGGTAGT AAAGGAGGGCAG UvrASchu4Dn1 5’GGAGAATTCCGCCATGCTCAACATCTCAAAGATG For FnPErmCSeq: pET15bUniUp: 5’ TGCATTAGGAAGCAGCCCAGTAGT pET15bUniDn: 5’ TTCCTTTCGGGCTTTGTTAGCAGC 3.12. Set up following reaction for UvrAUpSeq: 10 X KOD XL Buffer dNTP 2mM UvrAUp 2uM UvrADn 2uM U112 Chromosomal DNA 10 ng/ul KOD XL DNA polymerase dH2O 5.0 ul 5.0 ul 5.0 ul 5.0 ul 5.0 ul 0.4 ul 24.6 ul and UvrADnSeq: 10 X KOD XL Buffer 5.0 ul dNTP 2mM 5.0 ul UvrAUp1 2uM 5.0 ul UvrADn1 2uM 5.0 ul U112 Chromosomal DNA 10 ng/ul 5.0 ul KOD XL DNA polymerase 0.4 ul dH2O 24.6 ul at 94C 2’ then 94C 30”, 55C 30”, 72C 30” for 35 cycles, and at 72C for 10’. 19 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 3.13 Gel picture was shown as below: 1 2 3 4 5 6 Lane 1-3 UvrAUpSeq; Lane 4-6 UvrADnSeq. 3.14 For overlapping PCR, set up: Set up: 10 X KOD XL Buffer 5.0 ul dNTP 2mM 5.0 ul UvrAUp 2uM 5.0 ul UvrADn1 2uM 5.0 ul UvrAUpSeq 3.0 ul FpErmCSeq 3.0 ul UvrADnSeq 3.0 ul KOD XL DNA polymerase 0.4 ul dH2O 24.6 ul at 94C 2’, then 94C 30”, 55C 30”, 72C 3’ for 35 cycles, and 72C 10’. 3.15 The gel picture was shown for overlapping PCR as below. 1 2 3 4 Lane 1-4: UvrAUpSeqFpErmCSeqUvrADnSeq 20 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 3.16. The fragment from step 3.15 was inserted into pGEM-T. White colonies from Amp/X-gal plate were grown up. Plasmids were cut with EcoRI. 6 5 4 3 2 1 Lane 1-6: pGEM-TUvrAErmC plasmid The plasmid No.1 was sequenced and is correct. The plasmid was frozen away and designated as pKEK952. pKEK952 is the uvrA::ermC deletion plasmid. 3.17 pKEK952 was cryotransformed into U112, and plated onto TSA/ErmC plate. Colony PCR was performed to check mutant. 3.18 The gel picture was shown as below: 1 2 3 4 Lane 1: Negative control Lane 2: U112 Lane 3: pKEK952 Lane 4: Mutant 3.19. PCR fragments were purified from step 3.18, and were cut with BamHI. The mutant can be cut into two fragment as only it has BamHI site. The gel picture was shown as below: 21 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 1 2 3 Lane 1: U112 Lane 2: pKEK952 Lane 3: Mutant. 3.20 The PCR fragment from step 3.19 was sequenced, and is correct. The mutant was frozen away, and designated as KKF 72. KKF72 is the uvrA::ermC mutant in U112. 4. Significant decisions made or pending None 5. Problems or concerns and strategies to address No problems so far. 6. Deliverables completed pKEK951(UvrB deletion plasmid), KKF71( UvrB deletion mutant in U112);pKEK952(UvrA deletion plasmid), KKF72(UvrA deletion mutant in U112); pKEK887(Francisella promoter controlled ErmC plasmid). 7. Quality of performance Complete. 8. Percentage completed 90% of scientific work completed on the milestone 9. Work plan for upcoming month The double mutation uvrA, uvrB in U112 will be reported within this milestone in the coming month, though it is an addition to this milestone. 10. Anticipated travel None. 11. Upcoming Contract Authorization (COA) for subcontractors None. Milestone 40 Milestone description: Phenotyping of Ft novicida mutants; Measure degree of attenuation of uvr mutants in macrophages and in mice Institution: Cerus 1. Date started: 3/2/2006 2. Date completed: pending 3. Work performed and progress including data and preliminary conclusions 22 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Phenotyping of the F. tularensis ssp. novicida U112 uvrA strain from Dr. Karl Klose at UTSA is ongoing. We previously demonstrated that the uvrB mutant has no detectable growth defect in Chamberlain’s medium or in J774 macrophages. In order to determine whether uvrA has a growth defect compared to uvrB, or U112, the ability to replicate within J774 murine macrophages was measured. All three strains (uvrA, uvrB, U112) were able to replicate at the same rate (with a doubling time of approximately 2h). The data confirm our earlier finding that the deletion of uvrB does not render the bacteria more sensitive to macrophage-mediated killing, and suggest that the nucleotide excision repair pathway is not required for replication in macrophages. Intracellular Growth Curve in J774 CFU/ml 1.00E+08 1.00E+06 U112 U112 uvrA dl 1.00E+04 U112 uvrB dl 1.00E+02 NB920-103 1.00E+00 0 10 20 30 hours post infection 4. Significant decisions made or pending We have selected Chamberlain’s Defined Medium (CDM) and Cystine Heart Agar with Hemoglobin (CHAH) as liquid and plate medias for cultivation and enumeration of Ft novicida. 5. Problems or concerns and strategies to address Abrogation of the nucleotide excision repair pathway through uvrA or uvrB deletions does not appear to result in a macrophage growth defect. If this trend persists in animals, it suggests that a secondary attenuating mutation would be required if the SchuS4 strain were to be used as the vaccine background (since LVS is already attenuated in humans it does not require a secondary attenuating mutation). We will be screening attenuated Ft novicida mutants that also have uvr mutations for immunogenicity in milestone 43, with the ultimate goal of selecting an attenuating mutation to construct in SchuS4. We will also test a double mutant uvrA, uvrB. 6. Deliverables completed None 7. Quality of performance Good progress 8. Percentage completed 20% 9. Work plan for upcoming month 23 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble In the upcoming months, the degree of virulence of U112, uvrA and uvrB mutants will be determined in Balb/c mice using a subcutaneous model of infection. Our initial experiment will be designed to determine the appropriate LD50 dose range of each strain ranging from 1x10 2 to 1x108. 10. Anticipated travel Dr. Skoble, Dr. Bahjat, and Mr. Kim will be traveling to Albuquerque on September 26 to attend the UNM TVDC annual conference. 11. Upcoming Contract Authorization (COA) for subcontractors None Milestone 41 Milestone description: Optimization of psoralen treatment and characterization of KBMA F. novicida; Determine the amount of S-59 and UVA required to inactivate uvr mutants, determine extent of metabolic activity of uvr mutants after S-59 and UVA inactivation, determine the level of virulence attenuation of KBMA uvr strains in mice Institution: Cerus 1. Date started: 3/2/06 2. Date completed: pending 3. Work performed and progress including data and preliminary conclusions We previously identified the minimum concentration of S-59 required to inactivate wild type Ft novicida U112 was 20M and the minimum concentration required to inactivate Ftn uvrB was 5M when exposed to 6.5 J/cm 2 UVA. We found that complete inactivation was reproducibly achieved at 4 J/cm 2. Surprisingly, we found that both U112 and uvrB maintained metabolic activity for up to 12 hours. This month we have characterized the sensitivity of the Ftn uvrA strain to photochemical inactivation. 1) Two experiments were performed to determine the appropriate dose of S-59 required for complete inactivation of the uvrA mutant. In the first experiment complete inactivation was not achieved at 1, 2, 5, or 10uM but was achieved at 20 uM. In the second experiment more concentrations between 10 and 20 uM were tested. Complete inactivation was not achieved at 1, 2, 5, 10, or 12uM but was achieved at 15, 18, and 20uM. Thus, 15µM was the minimum concentration of S-59 required to achieve complete inactivation of Ftn uvrA using 4J/cm2 UVA. This is more S-59 than required for complete inactivation of the uvrB mutant and less than for U112. cfu/ml S-59 + 4J/Cm2 Ftn uvrA Kill Curve 1.0×10 11 1.0×10 10 1.0×10 09 1.0×10 08 1.0×10 07 1.0×10 06 1.0×10 05 1.0×10 04 1.0×10 03 1.0×10 02 1.0×10 01 1.0×10 00 uvrA -008 uvrA -022 NB947-008 NB947-022 0 5 10 15 20 [S59] uM 24 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 2) Knowing that 15uM S-59 and 4 J/cm2 allowed for complete inactivation we next determined the minimum amount of UVA light required for inactivation of the uvrA mutant. Below are data from one representative experiment of two in which the raw CFU data from three replicate plates are presented. We determined that complete inactivation of Ftn uvrA can be achieved with 15µM S-59 using 4 or 6.5 J/cm 2 UVA, but that 2J/cm2 provides inconsistent inactivation. This is similar to what was seen with the uvrB mutant. Plate Well ID B1 B2 B3 B4 B5 B6 C1 C2 C3 C4 C5 C6 A6 [S-59] 20uM 20uM 20uM 18uM 18uM 18uM 15uM 15uM 15uM 0uM 0uM 0uM 0uM UVA 2 J/cm2 4 J/cm2 6.5 J/cm2 2 J/cm2 4 J/cm2 6.5 J/cm2 2 J/cm2 4 J/cm2 6.5 J/cm2 2 J/cm2 4 J/cm2 6.5 J/cm2 0 J/cm3 Colony Count 1:10 Dilution 0,0,0 0,0,0 0,0,0 0,0,0 0,0,0 0,0,0 0,0,0 0,0,0 0,0,0 Not Done Not Done Not Done Not Done Neat 0,0,0 0,0,0 0,0,0 0,1,0 0,0,0 0,0,0 0,2,0 0,0,0 0,0,0 Not Done Not Done Not Done Not Done 10-7 Dilution Not Done Not Done Not Done Not Done Not Done Not Done Not Done Not Done Not Done Not Done Not Done Not Done 286,302,347 CFU/mL 0.00E+00 0.00E+00 0.00E+00 3.30E+00 0.00E+00 0.00E+00 6.60E+00 0.00E+00 0.00E+00 N/A N/A N/A 3.12E+10 (NB 948-039) 3) The metabolic activity of Ftn uvrA strain was determined when inactivated with 0, 15, 18, and 20 uM S-59 and using 4 J/Cm 2 UVA. The metabolic activity of the cultures was determined using Cell Titer 96 assay using a series of 3-fold dilutions of particles ranging from 1x 109 to 3x107. Below are representative data from the nominal 1e8 group. NB 948-039 Ft novicida uvrA Nominal 1.1E8cfu/mL 2.0 Well B2, 20uM S59, 4J/cm2 UVA 1.8 1.6 Well B5, 18uM S59, 4J/cm2 UVA OD490 1.4 1.2 Well C2, 15uM S59, 4J/cm2 UVA 1.0 0.8 0.6 Well C5, 0uM S59, 4J/cm2 UVA 0.4 0.2 0.0 0 2 4 6 Time, hrs 8 10 12 Well-A6 0uM S59, NO UVA In this experiment the groups that received no UVA or no S-59 (and were therefore live and capable of replication) demonstrated a higher degree of metabolic activity than the groups that were killed with S-59 and UVA. All three photochemically inactivated cultures demonstrated a reduced degree of metabolic activity that was indistinguishable from each other, but persisted for greater than 12 hours. 4) The effect of decreasing UVA dose on metabolic activity was also assessed. Below are curves generated using 15uM S-59 and 0, 2, 4, or 6.5 J/cm2 UVA. There was no significant 25 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble increase in the metabolic activity when UVA dose was decreased from 6.5 to 4 or 2 J/cm 2. All experiments were repeated with similar results not shown (NB948-058). Ft novicida uvrA Nominal 1.1E8cfu/mL 2.0 Well C1, 15uM S-59, 2J/cm2 UVA 1.8 Well C2, 15uM S-59, 4J/cm2 UVA 1.6 1.4 Well C3, 15uM S-59, 6.5J/cm2 UVA OD490 1.2 1.0 0.8 Well C5, 0uM S59, 4J/cm2 UVA 0.6 0.4 Well-A6 0uM S59, NO UVA 0.2 0.0 0 2 4 6 8 10 12 Time, hrs NB948-039 5) The difference in metabolic activity between KBMA Ftn uvrA and Heat killed Ftn uvrA was assessed using the Cell Titer 96 assay. The data generated with 3.3 x 108 initial input bacteria are shown below. While the live bacteria (UVA treated without S-59) had a higher degree of metabolic activity than the KBMA bacteria (treated with 15µM S-59 and 4 J/cm2 UVA), the heat killed bacteria had no measurable metabolic activity. Ft novicida uvrA 15uM S-59 4 J/cm2 Nominal 3.33e8 cfu/mL Well C2, 15uM S-59, 4J/cm2 UVA 3.5 3.0 Well C5, 0uM S59, 4J/cm2 UVA 2.5 OD490 2.0 1.5 Well-A1 0uM S59, NO UVA Heat Killed 1.0 0.5 0.0 -0.5 0.0 2.0 4.0 6.0 8.0 10.0 12.0 Time, hrs Conclusions: The uvrA mutant is slightly more sensitive to photochemical inactivation than the WT strain, but is slightly less sensitive than the uvrB mutant. The fact that the inactivation profiles are not identical between the two mutant strains suggests that perhaps there is potential for a compound effect of combining the two mutations. However, all 26 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble photochemically inactivated Ft novicida demonstrate significant metabolic activity for greater than 12 hours and display a vastly higher degree of metabolic activity than heat killed bacteria. This might be enough metabolic activity to maintain immunogenicity. 4. Significant decisions made or pending The minimum S-59 dose required for inactivation of U112 is 20 µM, the minimum S-59 concentration required for inactivation of Ftn uvrB is 5 µM and for Ftn uvrA is 15 µM. The minimum UVA dose required to achieve complete inactivation is 4 J/Cm 2. 5. Problems or concerns and strategies to address We previously have determined the concentration of S-59 required to inactivate Ftn uvrB is only one ¼ the concentration required to inactive the wild-type U112 strain. Currently, we have determined that the concentration of S-59 required to inactivate Ftn uvrA is ¾ the concentration required to inactive the wild type U112 strain. We have determined that the level of metabolic activity between Ftn U112 and uvrB or uvrA mutant strains is indistinguishable. This difference is less than we have observed for other organisms. It is still possible that the Ftn uvrA+uvrB strain will be more sensitive to photochemical inactivation, and hence may display a higher degree of metabolic activity. The uvrA+ uvrB double mutant will be evaluated as soon as it is received from UTSA . 6. Deliverables completed None 7. Quality of performance Good progress 8. Percentage completed 25% of scientific work completed on the milestone 9. Work plan for upcoming month We have determined the minimum concentration of S-59 required for complete inactivation of U112, uvrB and uvrA strains (20M, 5 µM and 15M respectively). We have determined that the lowest dose of UVA that results in reproducible complete killing (4 J/cm 2). We have determined that the U112 and uvrB and uvrA strains have comparable metabolic activity when inactivated. We will evaluate the sensitivity of the uvrA+ uvrB double mutant to photochemical inactivation will be as soon as it is received from UTSA . and we will compare the metabolic activity of the uvrA+ uvrB double mutant to the WT, uvrA, and uvrB mutant strains. We will begin to scale up the inactivation process. Additionally, a storage formulation will be developed for KBMA strains. 10. Anticipated travel Dr. Skoble, Dr. Bahjat, and Mr. Kim will be traveling to Albuquerque on September 26 to attend the UNM TVDC annual conference. 11. Upcoming Contract Authorization (COA) for subcontractors None Milestone 43 Milestone description: Create uvrA or uvrB mutants in LVS Institution: UTSA 1. Date started: 5/01/2006 2. Date completed: In progress 3. Work performed and progress including data and preliminary conclusions 27 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble 3.1 The pKEK1028 (pUCUvrBLVSUpSeq) and the pKEK1029 (pUCUvrBLVSDnSeq) plasmids were digested individually with BamHI ; then subsequently, these were digested with EcoRI enzyme to completion. 3.2 The plasmids above were run on a 1% agarose gel. The pKEK1029 plasmid was linearized with the enzymes mentioned and this was isolated from the gel using the Qiagen Extraction Kit mentioned in an earlier report. 3.3 The pKEK1028 generated two fragment sizes on the gel and these represented the plasmid (pUC19) as the larger band and the smaller band represented the 5´ end of the UvrBLVS sequence which was gel isolated with the extraction kit mentioned in 3.2. 3.4 Each of the isolates in 3.2 and 3.3 were used in a ligation reaction to generate a plasmid that contains the entire UvrBLVS deletion. Once the ligation reaction was complete this was phenol: chloroform extracted; ethanol precipitated; and used to transform into Top10 competent cells. 3.5 Ten plasmid preparations were made from some of the resulting transformants in 3.4. These where digested with EcoRI and compared to the parent vector (KEK1029). The correct construct yielded about a 1700 bp shift compared to the parent vector. Two candidates looked correct therefore, these were both digested with EcoRI and BamHI together to yield two bands on the gel. The smaller band represents the 5’ end of the UvrB fragment which looked correct for both clones. 3.6 A larger plasmid preparation was made from one of the potential candidates above to send for sequencing. 3.7 In the meantime, started to prepare the kanamycin fragment that we want to insert in the middle to the deletion to help in selecting for the desired UvrB deletion in the LVS strain. Will use KEK964 (pUC118 containing the Francisella promoter (Ft) in front of the kanamycin gene) to remove the fragment Ft+kanamycin gene by digestion with Bgl II then BamHI. All the data were documented in page 58-61, TVD UTSA notebook #2. 4.Significant decisions made or pending None 5. Problems or concerns and strategies to address None. 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed Approximate 38% scientific work completed on the milestone 9. Work plan for upcoming month 3.1 Prepare Ft+Kanamycin fragment to use in ligation reaction with confirmed UvrBLVS deletion sequence (plasmid in step 3.6). 3.2 Perform transformation of Top10 cells with this ligation and screen clones by digestion. 3.3 Once the correct construct with the UvrB deletion along with the Ft+Kan fragment is confirmed will proceed with crytransformation of LVS to generate the desired mutant. 10. Anticipated travel None. 11. Upcoming Contract Authorization (COA) for subcontractors None. 28 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Milestone 46 Milestone description: Scale up of KBMA LVS vaccine production; Optimize large–scale Francisella tularensis culture conditions, Establish 3L culture scale purification conditions, Optimize 3L scale photochemical inactivation process, Verify protective immunogenicity of vaccine candidates produced by optimized large-scale process Institution: Cerus 1. Date started: 3/2/2006 2. Date completed: pending 3. Work performed and progress including data and preliminary conclusions We have demonstrated previously that LVS grows robustly in Chamberlain’s defined medium (CDM) and that WT Ft novicida and Cerus’ strain of LVS are inactivated with similar concentrations of S-59. We have received our APHIS permit from USDA for import of LVS and Ft novicida (permit # 54834) and have received requested vials of DVC LVS. 1) Working cell banks of DVC lot# 16 LVS were prepared by growing cultures in CMD for 36 hours and were harvested at an OD of 1.0 and stored at –80 oC. 2) We attempted to calculate the ratio of Blue/Gray variants in the DVC material and compare to a single passage in CDM, but colonies failed to grow on cysteine peptone agar plates (provided by UNM from PML Microbiologicals). The expected number of colonies grew when the same dilutions were plated on CHAH plates. These data suggest that the cysteine peptone agar does not support robust growth of LVS, and therefore represent a selective growth condition and would be inappropriate for the quantification of colony phenotype. We will confer with other TVDC members about a standard assay we can all use for determining the LPS phase variation. 3) We have performed a S-59 dose-titration for photochemical inactivation of DVC LVS using the Cerus_Ft_protocol_005 to determine the minimum concentration of S-59 required for complete inactivation (focusing on concentrations between 1 and 50 µM). Complete inactivation of DVC LVS was achieved at all concentrations of S-59 greater than or equal to 5uM. This is the same concentration of S-59 required for inactivation of the uvrB mutant of Ftn. This is likely due to differences in sensitivity between the holarctia and the novicida subspecies. The sensitivity of the uvr mutants of LVS will be determined when the mutants are constructed. 29 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble NB 934-080 DVC Ft LVS Kill Curve (6-Well Format) 8.00E+01 7.00E+01 Initial Titer = 2.43e9 6.00E+01 cfu/mL 5.00E+01 4.00E+01 Series 1 3.00E+01 2.00E+01 1.00E+01 0.00E+00 0 5 10 15 20 25 30 35 40 45 50 [S-59] (uM) 4. Significant decisions made or pending We have selected Chamberlain’s Defined Medium (CDM) and Cystine Heart Agar with Hemoglobin (CHAH) as liquid and plate medias for cultivation and enumeration of LVS. We have switched from glycerol to DMSO for cryopreservation of stocks. We have determined the minimum concentration of S-59 psoralen required for complete inactivation is 5uM. . 5. Problems or concerns and strategies to address We do not know why LVS failed to grow in CDM in the fermentor but grows well in CDM in shaker flasks. In the future we will keep the media in a dark environment prior to cultivation as the media appears to be light sensitive. 6. Deliverables completed None 7. Quality of performance fair progress 8. Percentage completed 13% of scientific work completed on the milestone 9. Work plan for upcoming month We will repeat the DVC LVS S-59 titration with more concentrations between 0 and 5 uM. We will modify the large-scale propagation procedure so that the inoculum going into the fermentor is in early stationary phase rather than late stationary phase and attempt 3L-scale propagation using media stored in the dark. During the cultivation we will monitor the pH, dissolved oxygen concentration, and optical density of the bacteria and determine the number of colony forming units at various time points. 10. Anticipated travel Dr. Skoble, Dr. Bahjat, and Mr. Kim will be traveling to Albuquerque on September 26 to attend the UNM TVDC annual conference. 11. Upcoming Contract Authorization (COA) for subcontractors None 30 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Milestone 49 Milestone description: Construct single mutants in F. tularensis subsp. tularensis (SCHU S4) (iglC, pdpD, iglD, iglA, iglB) 49.1: Construct iglC F. tularensis subsp. tularensis (SCHU S4) 49.2: Construct pdpD F. tularensis subsp. tularensis (SCHU S4), Construct iglD F. tularensis subsp. tularensis (SCHU S4) 49.3: Construct iglA F. tularensis subsp. tularensis (SCHU S4), Construct iglB F. tularensis subsp. tularensis (SCHU S4) Institution: UTSA 1. Date started: April 1, 2006 2. Date completed: in progress 3. Work performed and progress including data and preliminary conclusions I. Cloning of igLC: a) Prepared10 more mini plasmid preparations from the transformation resulting from the ligation reaction of the IgLC deletion PCR product and the pUC118 KEK964 construct. Ran diagnostic digestions with NdeI and EcoRI; there were no promising candidates. b) Therefore, decided to try a lower copy number plasmid KEK229 to clone in the igLC deletion fragment. This plasmid is essentially pCVD442 containing the Multiple cloning site from pwsk30 vector instead of it’s own (see figure 1). It is imperative that we clone this deletion into a E.coli strain (via a plasmid) in order to continue our work in getting the deletion in Schu4. The igLC sequence seems to be toxic to these strains which makes it very unstable . Using a lower copy number plasmid may help offset this instability and allow us to clone this deletion in our E. coli strain via the plasmid. Figure 1 The bold print on the multiple cloning site bars indicate these restriction endonuclease sites are not unique. 31 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble c) The pCVD442 was created by Ried and Collmer (Gene 57:239-46, 1987) and contains the following features: i. a pir dependent origin of replication from R6K ii. a bla gene encoding resistance to ampicillin iii. a mob gene region allowing efficient transfer by conjugation from strains containing the tra locus iv. the sacB gene conferring sensitivity to sucrose in Gram-negative bacteria d. The plasmid containing the IgLC deletion with no resistance marker (KEK906will use the 3000 bp NotI/Sal I fragment) and KEK229 were both digested with Not I then subsequently with Sal I. Both were then run on an agarose gel and each were gel purified as previously described. The resulting isolates were used in ligation reaction. e. Transformation was performed with SM10λpir competent cells using the cleaned up ligation (i.e. phenol:chloroform extracted then ethanol precipitated) generated in d. The first attempt did not yield any colonies therefore, prepare fresh competent cells of SM10λpir and DH5αλpir. In addition, prepared fresh digestions and isolations of the two plasmids in d f. Prepared a new ligation reaction with new isolates and performed another transformation using both bacteria strains above, independently. g. Only two colonies resulted in the DH5αλpir strain; these were screened and found to be only re-ligation products (i.e. the original vector KEK229 with no insert). h. The SM10λpir transformation yielded 15 colonies; however, the background (i.e. religation plate was high, about 9 colonies). Have not been screened yet and will be screened next month. i. Data located in TVD UTSA Notebook 3, page 22-24. II. Experiments to generate deletions in Schu4: We are working with the MglA gene to show that we can create a deletion in SCHU S4; it is a single copy gene in this organism and that is easier to screen for than the genes we will be deleting in the project. We have a construct in hand which contains over 1000 bp homology at the 5´ and 3´ ends of this gene therefore, it is ideal to use in the development of deletion protocol. a. Using a plasmid (KEK1023) containing about 1200 bp homologous flanking regions of a gene called MglA; performed an electro-transformation into Schu4. b. This plasmid is pUC118 containing the F. tularensis promoter driving the expression of the kanamycin gene which encodes resistance to kanamycin. Immediately, downstream of this fragment is the MglA deletion gene (about 2400 bp fragment). We hoped that the homology would be long enough to allow recombination with the Schu4 chromosome as the plasmid enters the cell. c. The selection of the transformants were on 50 ug/ml (final concentration) kanamycin (Kan) Tryptic Soy Agar (TSA +++) plates containing various supplements and described earlier. Also, a negative control (i.e. Schu4 with no plasmid used in electroporation) was also plated on these plates to follow any spontaneous kanamycin resistant cells that may occur during our five day 37° C incubation that was used following the electroporation. d. After five days did not see any spontaneous resistant cells on the negative control plates. However, only five clones resulted from the electroporation with the KEK1023. e. Chromosomal preparations were made of these “parent” transformants to check for recombination into the Schu4 chromosome. Used primer set MglAB H3 and MglAB BHI to look for the deletion. If the plasmid got cross-over with Schu4 you will see two bands; one size will equal what you would see in wild type and the other will reflect the deletion size. According to DNA sequence information on Schu4 there is only one MglA gene in Schu4 therefore, if the deletion is complete then we should be left with only one product with this oligo set. (See figure 2) 32 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble f. These transformants were passaged onto non-selective plates to try and get the plasmid out of the chromosome. The single colonies were struck on these TSA+++ plates to generate single colonies and these were patched on Kan plates and TSA+++ nonselective to look for sensitivity to Kan. Each cycle of passage would be taken from the previously passaged plate to create the next cycled plate. g. Performed four passages by plates and never generated a sensitive Kan colony therefore, did four passages in liquid culture, made dilution from each passage and spread on non-selective TSA+++ plates. These were patched onto Kan and TSA +++ plates to look for Kan sensitive clones. No Kan sensitive clones resulted but I did make chromosomal preparations from some representative singles generated from each passage to check by PCR to see if the deletion was in the chromosome with the plasmid or if the deletion was lost and the plasmid was left in the chromosome (very unlikely). (See figure 2) Figure 2: a. Below represents one of the PCR screens run with the five clones generated from KEK1023 electrotransformed into Schu4. The Legend identifies the strain, clone, or plasmid used as the template for the PCR reaction. The oligo set used here is MgLAB H3 & MgLAB BHI. Those clones that do not have “TK” in their name are single colonies from one of the passages made in the experiment. The Schu4 WT product is the reference for native MgLA gene product. The KEK1023 is the plasmid containing the MgLA gene deletion that should have entered the chromosome therefore, can serve as the deletion product size. The KKF34 is the MgLA deletion in F.novicida strain, this is just to verify primer set is working correctly. Data located in TVD UTSA Notebook 3, page 33. The deletion sequence did cross-over into the Schu4 chromosome;however, since the plasmid also has entered the chromosome the PCR profile will give both the wild-type (WT) and the deletion size band (it is not a completed deletion). The control WT strains on the gel have no plasmid and the profile is yielding only the WT gene size as expected. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Legend Figure 2a: 1. 1 Kb ladder 2. Schu4 WT 3. KKF34 U112dMgLA 4. KEK1023 5. 3TKA 6. 3AI 7. 3TKB 8. Schu4 WT 9. TypeB WT 10. 3BI 11. 3TKC 12. 3TKD 13. 3TKE 14. 3DI b. This represents another PCR screen using the indicated strain, clone or plasmid used as a template. The oligo set here used in the reaction are PUCori XhoI and PUCori EcoRI. The set is used to confirm whether or not a second recombination has occurred causing the plasmid to be lost from the chromosome. The Schu4 WT should not have the PUC origin; as well as, the KKF34 strain, since the deletion was made with a PCR product with no plasmid. The KEK1023 is the vector used in experiment and yields the correct PUC origin PCR product size that should result from the oligos used in this screen. The passaged clones used in this set are later single isolates which still remain Kan resistant. We did not generate any Kan sensitive mutants however, I checked a few passage single clones to verify primer set is working properly. Data located in TVD UTSA Notebook 3, page 35. Since we did not generate any Kan sensitive 33 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble mutants, I checked a few passage single clones to verify primer set is working properly. This is checking for the plasmid’s origin of replication sequence; a oligo set checking for the kanamycin gene itself was also used and it yielded a PCR product as expected (data not shown). 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Legend Figure 2b: 15. 1 Kb ladder 16. Schu4 WT 17. KKF34 U112DMgLA 18. KEK1023 19. 3TKA 20. 3A6 21. 3TKB 22. 3B4 23. 3TKC 24. 3C4 25. 3TKD 26. 3D7 27. 3D8 28. 3D9 . h. The deletion seems to remain in the chromosome but we are unable to get the second recombination to occur to remove the plasmid. It may be possible if we use the SacB gene expressed from the Ft promoter we may be able to force the second recombination and loose the plasmid. i. Data located in TVD UTSA Notebook 3, page 31-36. 4. Significant decisions made or pending None 5. Problems or concerns and strategies to address We are using various approaches to deleting the IglC gene in SCHU S4 and in summary our stratagies are: 1. The first plasmid used to clone in the entire igLC deletion (about 3000 bp fragment) was in KEK903 which was the original mating plasmid proposed to use in creating deletions in Schu S4. We were able to get a correct clone from our transformations. 2. Secondly, we tried using pKEK999 which is a pUCvector containing a F. tularensis promoter driving the Kanamycin gene expression. This was going to be used to create mutants in Schu S4 by way of cryotransformation or electroporation instead of a mating method. We were unable to get the deletion in this plasmid. 3. Currently, we will attempt to clone this deletion in a pKEK229 which is a mating vector used in our lab and is a low copy number plasmid. More detail on this plasmid is given in the text of this report. 6. Deliverables completed None 7. Quality of performance Good 8. Percentage completed 17% 9. Work plan for upcoming month a. Prepare more mini plasmid preparations from the transformation resulting from KEK229+iglC deletion sequence in section I.h. above. We are hopeful that this set will yield the correct construct containing the longer iglC deletion to use in making the Schu4 igLC deletion. b. Once we verify the correct construct, I will try cryotransformation and electrotransformation to attempt to generate the IglC deletion in Schu4. 34 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble c. Will work also in cloning in the SacB gene in KEK1023 to check if this would be helpful in pushing the second recombination event needed to generate a mutation in Schu4. d. Order more supplies as needed 10. Anticipated travel None 11. Upcoming Contract Authorization (COA) for subcontractors None Milestone 50 Milestone description: Phenotyping and confirmation of single gene mutants; 50.1: phenotyping and immunologic characterization of Ft subsp. novicida uvrA or uvrB; LVS uvrA or uvrB, and Ft subsp. tularensis (SCHU S4) iglC strains, 50.2: phenotyping and immunologic characterization of Ft subsp. tularensis (SCHU S4) pdpD, iglD strains, Ft subsp. novicida uvrA or uvrB plus pdpD/iglA/iglB/iglC/iglD double mutant strains, 50.3: phenotyping and immunologic characterization of Ft subsp. tularensis (SCHU S4) iglA, iglB strains Institution: UTSA 1. Date started: 04/01/2006 2. Date completed: provide date when milestone is completed 3. Work performed and progress including data and preliminary conclusions a. These experiments were performed to verify and obtain basal level of information on respiratory cellular responses with Ft LVS and Ft novicida and will be used comparatively with the attenuated mutants that will be created by Dr. Klose. Mice (BALB/C 6-week-old females) were challenged intranasally with 10 LD50 of Ft LVS (20,000 CFU, live or UVinactivated) or Ft. novicida (Fn, 100 CFU, live or UV-inactivated). Lungs were collected from Ft-infected mice 24h-, 48h-, and 72h-post challenge (3 mice/time point/group), single cells were made and subjected to flow cytometric analysis to determine the infiltrating populations of antigen presenting cells-macrophages, dendritic cells, neutrophils and NK cells (Note Book #4 page:31-33, 34-36). Naive and PBS-treated mice were used as a base line control. In our July report, we have shown that populations of T cells and B cells did not differ among all examined groups and a similar phenotype analyses on spleen cells from those infected animals did not show any remarkable differences. Summary of the Flow Cytometric Analyses 1. Whereas there was minimal influx of APCs (Antigen Presenting Cells) at 24 hr with F. novicida challenge, UV-inactivation of the organism greatly increased the numbers of cells in the lung at this time point (Table 1, File name: 0806 infiltrate.doc). 2. At 48 hr, there was an increase in the cellular infiltration after challenge with F.novicida, while the response with the UV-inactivated organism has begun to wane. 3. By 72 hr the cellular response in the lungs of either F.novicida or UV-inactivated organism had dropped back to a baseline. In contrast, very minimal differences were observed with LVS and UV-inactivated LVS, presumably because of the attenuated nature of the strain. Together these results, suggest that F. novicida may be suppressing/interfering with the initial cellular response in the lung as early as 24 hr as evidenced by the differences in cellular infiltration between the replicating and killed organism. To our knowledge, although this phenomenon has been reported in the literature, there has been no substantial data to really show this effect in vivo. 35 of 36 Tularemia Vaccine Development Contract: Technical Report Period: 8/01/2006 to 8/31/2006 Due Date: 9/15/2006 and Prepared by: C. Rick Lyons, Barbara Griffith, Terry Wu, Bob Sherwood, Julie Wilder, Ed Barr, Mitch Magee, Kathryn Sykes, Stephen Johnston, Karl Klose, Justin Skoble Table 1. Cellular infiltration in lungs of mice upon pulmonary Francisella infection Lungs 24 hr Isotype CD11b 2.2 CD11c 1.2 CD59 1.2 Ly6 1.2 Naive 2.2 PBS 2.1 6 TOTAL UV-Fn 3.8 10.1 6.7 5.2 13.5 3.9 2.2 7.6 5.9 3.9 12.9 4.5 4.6 48 hr F nov 14.9 21 Lungs 15.1 44.1 PBS UV-Fn 10.5 3.6 7 12 1 6.5 5.1 13.4 4.7 2.8 15.9 24 hr Isotype CD11b 0.3 CD11c 0.1 CD59 0.3 Ly6 0.1 TOTAL Naive 6.4 PBS 4 2.1 8.8 21.3 F nov UV-Fn 3.4 5.4 5.7 6.6 6.3 8.2 7.9 1.9 2 4.6 3.8 4.8 7.1 6.6 42.4 16.8 PBS 16.5 48 hr LVS UV-LVS 5.2 2.8 4.8 4.3 2.8 3.2 1.9 1 7.1 5.7 18.5 72 hr F nov UV-LVS 6.6 2.1 4.2 3.6 2.1 2.9 1.2 1.3 0.5 1 5.9 7.5 2.7 6.2 15.1 19 7.4 24 72 hr LVS 12.3 PBS 25.3 14.3 PBS LVS UV-LVS 6.8 5.8 4.5 5.6 4 3 1.6 1.2 1.3 8 7.7 5.7 22 18.7 Mice (BALB/C 6-week-old females) were challenged intranasally with 10 LD 50 of Ft. subspecies novicida (100CFU, live or UV-inactivated) or Ft LVS (20,000 CFU, live or UV-inactivated). Lungs were collected from Ft-infected mice 24h-, 48h-, and 72h-postinfection (3 mice/time point/group), single cells were made and subjected to flow cytometry analysis to determine the population of inflammatory cells. CD11b, CD11c, CD59, and Ly6, are commonly used markers for macrophages, dendritic cells, NK cells, and neutrophils, respectively . 4. Significant decisions made or pending None 5. Problems or concerns and strategies to address None 6. Deliverables completed None 7. Quality of performance Good” 8. Percentage completed Provide approximate 15% of scientific work completed on the milestone 9. Work plan for upcoming month a. Receive Ft subsp. novicida uvrA strain from Dr. Klose for characterization b. Determine the LD50 of Ft subsp. novicida uvrA c. Measure intramacrophage (J774) survival of uvrA 10. Anticipated travel None 11. Upcoming Contract Authorization (COA) for subcontractors None 36 of 36 14.5