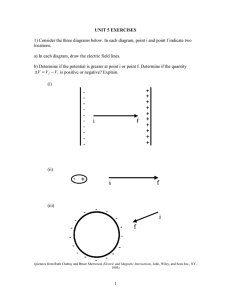
BIOLOGY 3400 Principles of Microbiology The
advertisement

The University of Lethbridge BIOLOGY 3400 Principles of Microbiology LABORATORY MANUAL Spring, 2016 Written by: L. A. Pacarynuk and H.C. Danyk Revised: December, 2015 TABLE OF CONTENTS Exercise: Page Biology 3400 Laboratory Schedule.................................................................................2 Biology 3400 Laboratory Expectations...........................................................................3 Grade Distribution..........................................................................................................4 Lab Book Preparation ………………………………………………………………………………………………..5 Guidelines for Safety Procedures...................................................................................7 Exercise 1 – Microscopy and Gram Staining: A Review ...............................................10 Exercise 2 – General Laboratory Principles and Biosafety...........................................13 Exercise 3 – Free-Living Nitrogen Fixation....................................................................15 Exercise 4 – Soil and Compost Microbial Ecology ........................................................23 Exercise 5 – Bacterial Reproduction.............................................................................33 Exercise 6 – Fermentation............................................................................................36 Exercise 7 – Virology.....................................................................................................41 Appendix 1 – The Compound Light Microscope...........................................................47 Appendix 2 – Preparation of Scientific Drawings.........................................................54 Appendix 3 – Staining……………………………..……………………………………………………………….56 Appendix 4 – Aseptic Technique..................................................................................60 Appendix 5 – The Cultivation of Bacteria.....................................................................65 Appendix 6 – Bacterial Observation.............................................................................71 Appendix 7 – Laboratory Reports.................................................................................72 Appendix 8 – Use of the Spectrophotometer...............................................................74 Appendix 9 – Media, Reagents, pH Indicators.............................................................76 1 BIOLOGY 3400 LAB SCHEDULE SPRING, 2016 Jan.7 No lab Jan. 12 Jan. 14 Introduction, Microscopy and Gram Staining: A Review General Lab Procedures, Biosafety, N-Fixation (inoculate broths) Jan. 19 General Lab Procedures, Biosafety – Complete; Soil/Compost Bacteria Enumeration – Begin Soil Bacteria Enumeration – Complete; Prepare Streak Plates; N-Fixation Jan. 21 Jan. 26 Jan. 28 Morphological Characterization of Unknown (Gram Stain); Subculture Unknown; N-Fixation Endospore/Capsule Stains; Unknown Identification – Develop Outline for Identification Feb. 2 Feb. 4 Unknown Identification; Catabolic Activity Unknown Identification; Catabolic Activity Feb. 9 Feb. 11 Unknown Identification: N-Fixation PCR Unknown Identification; N-Fixation Electrophoresis Feb. 16 Feb. 18 Reading Week Reading Week Feb. 23 Feb. 25 Bacterial Growth N-Fixation: Sequence Analysis Mar. 1 Mar. 3 Fermentation Part A: Yogurt Fermentation Part A: Yogurt - Complete Mar. 8 Mar. 10 Fermentation Part B: Yeast Fermentation Yeast Fermentation – sample and enumerate Mar. 15 Mar. 17 Virology (phage isolation); Yeast Fermentation - enumerate Virology (phage elution); Fermentation Part B: Yeast Fermentation Complete Mar. 22 Mar. 24 Virology (amplification) Virology (titre/host range) Mar. 29 Mar 31 Virology – Complete No lab Apr. 5 Apr. 7 Apr. 12 No lab No lab Lab Exam 2 Biology 3400 Laboratory Expectations: Biology 2000 is the prerequisite for Biology 3400. In turn, Biology 1010 and Biology 1020 serve as prerequisites for Biology 2000. Biology 3400 laboratories will build upon concepts introduced in these junior level courses. We will not spend laboratory time reviewing material covered in lectures and laboratories associated with these courses. If material/lab techniques are unfamiliar to you, it will be up to you to become proficient. The following list provides some guidance with respect to concepts and skills in which you should be proficient to be successful in microbiology labs this semester: Concepts: Prokaryotic cell structure (versus structure of eukaryotic cells) Prokaryotic cell division Bacterial DNA replication Polymerase chain reaction and gel electrophoresis Respiration and fermentation Catabolism Viruses; especially bacteriophage (lytic cycle) Skills: Aseptic technique Gram staining Culture inoculation (broths and plates) Appropriate handling of biologicals and chemicals Use of a dichotomous key Microscopy – dissecting and compound light microscopes, oil immersion Serial dilutions Keeping a lab book Scientific drawings Scientific method principles Critical thinking Data analysis and drawing appropriate conclusions Writing lab reports Searches of peer-reviewed literature 3 Laboratory Grade Distribution: The laboratory component of Biology 3400 is worth 50% of your course mark. It is distributed as follows: Skills Tests Assignments and/or Lab Reports Lab Books Lab Exam 8% 24% 8% (to be handed in multiple times) 10% Performance: Up to 10% of laboratory grade (5 marks out of 50) will be subtracted for poor laboratory performance. This includes (but is not limited to) failure to be prepared for the laboratory, missing lab notebook or lab manual, poor time management skills, improper handling and care of equipment such as microscopes and micropipettors, and unsafe practices such as not tying hair back, chewing gum, applying lipstick, eating, drinking, or chewing on pencils, and sloppy technique leading to poor results. As we are working with potential pathogens, students displaying improper or careless techniques will be asked to leave the lab and will have at least 5% of their laboratory grade deducted immediately. Missing a lab for which there is a skills test or assignment requires documentation. Upon presentation of this documentation, you will either have to complete the assignment or skills test as soon as possible or, if this is not possible, your lab grade will be recalculated. The lab books will be collected and graded several times during the semester. Although most exercises are completed as groups, the lab books are to be completed individually, and must represent individual effort. The following page provides you with tips on how to construct your books. Unannounced skills tests will be given during the semester. Students are expected to work independently on some technical aspect of microbiology and will be graded based on their techniques and their results. As proficiency in microbiological techniques is considered an essential component of the course, students are only permitted three lab period absences (you do not require any documentation). Missing more than three labs will result in a grade of 0 being assigned for the lab (at this point, it is recommended that students consult with Arts and Science Advising for the option of completing the laboratory the following year). Students are still responsible for the material missed (and their assignments, lab reports etc. will be graded as such). There are no make-up laboratories. Late Reports will be penalized as follows: less than 24 hours late, –10%; between 24-48 hours late, -25%, between 48-72 hours late, -50%, after 72 hours, no marks will be given. 4 Extensions for the lab reports and assignments will only be granted for situations involving prolonged illness (documentation is required). Preparation of a Lab Book: Your lab book provides you with a detailed record of your experiments performed. This record proves invaluable when preparing manuscripts for publication, or, more immediately, when preparing lab reports. This lab book, as with all of the reports and assignments is an individual effort. Choice of Lab Book Standard black lab books can be purchased from the bookstore but these are not required for this course. The only required features are: Pages are non-removable (no spiral bindings) All pages must be numbered in the top outer corner In General all entries must be made in blue or black ink (except drawings) date EVERY entry never remove a page or use white-out if an entry needs to be deleted, strike out the entry with a single straight line (the deleted entry must be readable) keep up to date, a lab book is meant to be filled out as the experiments are carried out and NOT after the fact record anything that may be useful to you when preparing your lab reports leave plenty of space throughout the lab book to add comments after the fact. Many exercises run over multiple periods – it is easier to organize your entries if you leave several blank pages for each exercise. Table of Contents Designate the first 2 pages as the Table of Contents record information and page numbers as you go Lab Entries For each lab be sure to include the following; 1. Objective 2. Method Summary do not rewrite the protocol from the lab manual highlight any specific changes to the lab protocol include times and dates for when work was performed record product and bacterial names and manufacturers used - enzymes, chemicals, equipment (micropipettors, baths) include incubation conditions for cultures and reaction 3. Observations & Results record any & all observations, this goes beyond number results include diagrams and any other form of raw data include calculations as appropriate 5 4. Conclusions did you achieve your objective? Why or why not? use your results to support your conclusions 5. Answer the thought questions at the end of the lab (as applicable) use reference citations as needed; these may be graded. 6 GUIDELINES FOR SAFETY PROCEDURES Students enrolled in laboratories in the Biological Sciences should be aware that there are risks of personal injury through accidents (fire, explosion, exposure to biohazardous materials, corrosive chemicals, fumes, cuts, etc). The guidelines outlined below are designed to: a) minimize the risk of injury by emphasizing safety precautions and b) clarify emergency procedures should an accident occur. EMERGENCY NUMBERS City Emergency Campus Emergency Campus Security Student Health Centre 911 (403) 329-2345 (403) 329-2603 (403) 329-2484 THE LABORATORY INSTRUCTOR MUST BE NOTIFIED AS SOON AS POSSIBLE AFTER THE INCIDENT OCCURS. EMERGENCY EQUIPMENT: Your lab instructor will indicate the location of the following items to you at the beginning of the first lab period. Closest emergency exit Closest emergency telephone and emergency phone numbers Closest fire alarm Fire extinguisher and explanation of use Safety showers and explanation of operation Eyewash facilities and explanation of operation First aid kit GENERAL SAFETY REGULATIONS: Eating and drinking is prohibited in the laboratory. Keep pencils, fingers and other objects away from your mouth. These measures are to ensure your safety and prevent accidental ingestion of chemicals or microorganisms. Personal protective wear is mandatory. Lab coats, safety glasses and closedtoed shoes must be worn at all times during lab exercises which involve potential for chemical or biological spills. Coats, knapsacks, briefcases, etc. are to be hung on the hooks provided, stowed in the cupboards beneath the countertops, or placed along a side designated by your instructor. Take only the absolute essentials needed to complete the exercise* with you to your laboratory bench. (* e.g. manual, pen or pencil) Mouth pipetting is NOT permitted; pipet pumps are provided and must be used. Always wash your hands prior to leaving the laboratory. Students are not allowed access to the central Biology Stores area for any reason. Consult your instructor if you require additional supplies. Report any equipment problems to instructor immediately. Do NOT attempt to fix any of the equipment that malfunctions during the course of the lab. Use caution when handling chemical solutions. Consult the lab instructor for instruction regarding the clean-up of corrosive or toxic chemicals. 7 Contain and wipe up any spills immediately and notify your lab instructor (see SPILLS below). Heed any special instructions outlined in the lab manual, those given by the instructor or those written on reagent bottles. Long hair must be restrained to prevent it from being caught in equipment, Bunsen burners, chemicals, etc. Dispose of broken glass, microscope slides, coverslips and pipets in the specially marked white and blue boxes. There will be NO disposal of glassware in the wastepaper baskets. You are responsible for leaving your lab bench clean and tidy. Glassware must be thoroughly rinsed and placed on paper toweling to dry. SPILLS: Spill of SOLUTION/CHEMICAL: While wearing gloves, wipe up the spill using paper towels and a sponge as indicated by the lab instructor. Spill of ACID/BASE/TOXIN: Contact instructor immediately. DO NOT TOUCH. BACTERIA SPILLS: If necessary, remove any contaminated clothing. Prevent anyone from going near the spill. Cover the spill with 10% bleach and leave for 10 minutes before wiping up. Discard paper towels in biohazard bag. Discard contaminated broken glass in designated biohazard sharps container. DISPOSAL: • Broken glass, microscope slides, coverslips and Pasteur pipets are placed in the upright white ‘broken glass’ cardboard boxes. NO PAPER, CHEMICAL, BIOLOGICAL OR BACTERIAL WASTE MATERIALS should be placed in this container • Petri plates, microfuge tubes, pipet tips should be placed in the orange biohazard bags. The material in this bag will be autoclaved prior to disposal. • Bacterial cultures in tubes or flasks should be placed in marked trays for autoclaving. • Liquid chemicals should be disposed of as indicated by the instructor. DO NOT dispose of residual solution in the regent bottles. In case of any uncertainty in disposal please consult the lab instructor. • Slides of bacteria should be placed in the trays filled with 10% bleach that are located at the ends of the laboratory benches. HEALTH CONCERNS: Students who have allergies, are pregnant, or who may have other health concerns should inform their lab instructor so that appropriate precautions may be taken where necessary. More information on Laboratory Safety is available on the Risk and Safety Services webpage: http://www.uleth.ca/risk-and-safety-services/content/lab-safety 8 STUDENT SAFETY ACKNOWLEDGEMENT This form must be completed, signed and submitted to the laboratory instructor before any laboratory work begins. I have read carefully and understand all of the safety requirements and procedures for working safely in the lab. I also agree to read all rules for specific exercises contained in the laboratory manual or laboratory handouts required for this course. I recognize that it is my responsibility to strictly follow them. I understand that I am required to wear personal protective equipment, such as safety glasses, lab coat, gloves etc., at all times when directed to do so in the laboratory. I further understand that I am permitted to work in the laboratory only under the supervision of a laboratory instructor. If I am unsure of the potential hazards related to lab procedures, I will discuss this with my instructor prior to undertaking the procedure in question. Name: ___________________________________________________ (please print) Student ID No.: _____________________________________________ Signature: ________________________________________________ Course: __________________________________________________ Date: ___________________________________________________ FOIP Notification: The personal information on this form is collected under the authority of the Post-secondary Learning Act (Alberta) and the Freedom of Information and Protection of Privacy Act (Alberta) for the purpose of administering the University’s laboratory safety program. For questions on the collection, use, and disclosure of this information, please contact the University’s FOIP Coordinator at 4401 University Drive West, Lethbridge, AB T1K 3M4; 403-332-4620; foip@uleth.ca. 9 EXERCISE 1 MICROSCOPY AND GRAM STAINING – A REVIEW Observing Bacteria Three fundamental properties of bacteria are size, shape and association. Bacteria generally occur in three shapes: coccus (round), bacillus (rod-shaped), and spirillum (spiral-shaped). Size of bacterial cells used in these labs varies from 0.5 m to 1.0 m in width and from 1.0 m to 5.0 m in length, although there are a range of sizes which bacteria demonstrate. Association refers to the organization of the numerous bacterial cells within a culture. Cells may occur singly with cells separating after division; showing random association. Cells may remain together after division for some interval resulting in the presence of pairs of cells. When cells remain together after more than a single division, clusters result. Cell divisions in a single plane result in chains of cells. If the plane of cell division of bacilli is longitudinal, a palisade results, resembling a picket fence. Both bacterial cell shape and association are usually constant for bacteria and hence, can be used for taxonomic identification. However, both properties may be influenced by culture condition and age. Further, some bacteria are quite variable in shape and association and this may also be diagnostic. Micrometry When studying bacteria or other microorganisms, it is often essential to evaluate the size of the organism. By tradition, the longest dimension (length) is generally stressed, although width is sometimes useful for identification or other study. Use of an Ocular Micrometer (Figure 1) An ocular micrometer can be used to measure the size of objects within the field of view. Unfortunately, the distance between the graduations of the ocular micrometer is an arbitrary measurement that only has meaning if the ocular micrometer is calibrated for the objective being used. 1. Place a micrometer slide on the stage and focus the scale using the 40x objective. 2. Turn the eyepiece until the graduations on the ocular scale are parallel with those on the micrometer slide scale and superimpose the micrometer scale. 3. Move the micrometer slide so that the first graduation on each scale coincides. 4. Look for another graduation on the ocular scale that exactly coincides with a graduation on the micrometer scale. 5. Count the number of graduations on the ocular scale and the number of graduations on the micrometer slide scale between and including the graduations that coincide. 10 6. Calibrate the ocular divisions for the 40x and the 100x objective lenses. Note that immersion oil is not necessary for calibration. Figure 1. Calibration of an ocular micrometer using a stage micrometer. The mark on the stage micrometer corresponding to 0.06 mm (60 m) is equal to 5 ocular divisions (o.d.) on the ocular micrometer. 1 ocular division equals 60 m/5 ocular divisions or 12 m. Once an ocular micrometer has been calibrated, objects may be measured in ocular divisions and this number converted to m using the conversion factor determined. Bacterial size is generally a highly heritable trait. Consequently, size is a key factor used in the identification of bacterial taxa. However, for some bacteria, cell size can be modified by nutritional factors such as culture media composition, environmental factors such as temperature, or other factors such as age. METHODS: 1. Use the diagram in Figure 1 to calibrate the 40x and the 100x objectives on your compound microscopes. Consult Appendix 1 if you need to review how to operate a compound light microscope. Record these values in your lab book as you will then use these values when measuring cells and structures for the rest of the lab. 2. Note: Do NOT use immersion oil when calibrating the 100x objective. This is the ONLY time during the term that you will not use immersion oil with this objective. 3. Prepare Gram stains of the cultures provided (consult Appendix 3 to review the procedure, if needed). 4. Prepare scientific drawings of organisms stained (consult Appendix 2 to review how to make scientific drawings). 11 THOUGHT QUESTIONS: 1. Why is it necessary to stain microorganisms before viewing them with the compound light microscope? 2. Explain how the Gram stain differentiates between Gram positive and Gram negative cells (your answer should address bacterial structure AND purpose of each of the components of the Gram stain). 12 EXERCISE 2 GENERAL LABORATORY PROCEDURES AND BIOSAFETY A primary feature of the microbiology laboratory is that living organisms are employed as part of the experiment. Most of the microorganisms are harmless; however, whether they are non-pathogenic or pathogenic, the microorganisms are treated with the same respect to ensure that personal safety in the laboratory is maintained. Careful attention to technique is essential at all times. Care must always be taken to prevent the contamination of the environment from the cultures used in the exercises and to prevent the possibility of the people working in the laboratory from becoming contaminated. Ensure that you have read over the guidelines on Safety, and those on Aseptic technique (Appendix 3). As well, you should be familiar with the contents of the University of Lethbridge Biosafety web site: http://www.uleth.ca/artsci/biological-sciences/bio-safety METHODS Part 1: General Laboratory Procedures A. Work individually to prepare a streak plate and a broth culture using the E. coli cultures provided. Refer to Appendix 3 as necessary. B. Work individually to prepare serial dilutions of the E. coli culture provided. 1. Prepare two spread plates on LB using 100 l of your 10-6 dilution for each. Place your plates on the appropriately labelled tray for incubation at 37 oC for 18 h. Record how you set up your serial dilutions in your lab book. 2. In the next lab period, count the number of colonies on your spread plates, and calculate the mean CFU/mL of original culture. Record this number on the board and in your lab book. 3. Calculate the mean CFU/mL (+/- SD) for your class, and compare your value to these. How successful were you at preparing serial dilutions? Part 2: Biosafety You will be provided with the following: Sterile swabs Sterile water Potato Dextrose Agar (PDA) plates and Luria Bertani (LB) plates Work in pairs to complete the following exercise: 1. Draw a line on the back of each plate to divide the plates in half. Label one half of the plate with the name of the surface to be tested. Label the other half of the plate as “after disinfection”. 2. Moisten the swabs provided with a small amount of sterile water. Brush the surface to be tested with the swab, and then use the swab to inoculate one-half of each of your two plates. 3. Disinfect the surface, moisten another swab, and repeat using the other half of both plates. Wrap the plates with parafilm. 13 4. Your plates will be incubated for 16-20 hours at 30oC, and then refrigerated at 4oC. During the next laboratory period, evaluate your plate results and record the number of colonies present on each half of both plates. Make observations of colony morphology. Thought Questions: (Use the Biosafety Web Site as a reference) 1. Were differences in colony morphology and number observed on the two types of media? Why? 2. Does disinfection of work surfaces completely eliminate all microbial organisms? What evidence do you have? 3. What is an MSDS and where can you find one? 4. In Canada, the Laboratory Centre for Disease Control has classified infectious agents into 4 Risk Groups using pathogenicity, virulence and mode of transmission (among others) as criteria. What do these terms mean? 5. What criteria would characterize an organism classified in Risk Group 1, 2 3 or 4? 6. There are many “Golden Rules” for Biosafety. Identify 4 common sense practices that will protect you in your microbiology labs. 14 EXERCISE 3 FREE-LIVING NITROGEN FIXATION PART A: ISOLATION OF FREE-LIVING NITROGEN FIXING MICROORGANISMS Nitrogen is an important component of amino acids, cell walls and other cofactors present in all cells. Nitrogen gas comprises greater than 75% of our atmosphere, but it is one of the most stable bonds in nature and is unavailable for use in this form. At one time early in the evolutionary history of life on earth, all cells may have had the ability to fix N2 gas into a more usable form (nitrate, nitrite or ammonia). Today however, only a few species of bacteria and archaea are capable of converting N2; all other organisms rely on N2-fixing prokaryotes for their fixed nitrogen requirements. M.W. Beijerinck, a Dutch microbiologist, successfully isolated free living nitrogen fixing bacteria in 1901. He inoculated soil samples into enrichment media containing glucose and mineral salts, but lacking any source of nitrogen other than atmospheric nitrogen. He observed cells that are today identified as members of the genus Azotobacter. Subsequently, other aerobic, free-living nitrogen fixing genera of bacteria have been isolated and identified, including Azomonas, Azospirillum and Beijerinckia. Nitrogen fixation occurs only when an enzyme called nitrogenase is present. The enzyme consists of two distinct proteins (i) dinitrogenase, which reacts with N2, and (ii) dinitrogenase reductase, which reduces nitrogen gas to ammonia. The dinitrogenase reductase component is irreversibly inactivated by the presence of oxygen. Several strategies have evolved to enable free-living, aerobic organisms like Azotobacter to fix nitrogen. Azotobacter has a very high respiratory rate, which is thought to prevent any stray oxygen from coming into contact with the nitrogenase enzyme. Additionally, freeliving nitrogen fixers often secrete copious amounts of slime which may prevent extra oxygen from entering the cells. There is also evidence suggesting that in the presence of oxygen nitrogenase can combine with a specific protein inside the cell that shields the oxygen sensitive site and prevents it from interacting from oxygen. When oxygen levels drop, nitrogenase can resume its activity. Over the course of the semester we will isolate free living nitrogen fixing bacteria from prairie soil, establish pure cultures and attempt to identify cultures using modern day molecular techniques. METHODS: For each lab: 5, 250 mL flasks containing N-free medium 10 plates N-free medium Balance, weigh boats and spatula N-free soil sample 15 1. Work in groups of 4 to inoculate your flasks. 2. Weigh out 1 g of the soil sample provided, and add it to an Erlenmeyer flask containing 100 mL of N-free medium. 3. Swirl gently to mix. Label the flask with your lab, bench number, and date. Make sure that the cap or foil is loosened sufficiently to allow air to enter the culture. Incubate for one week at 30oC. 4. After 7 days, remove the flask and look for the presence of a thin film of growth on the surface of the medium. Use a sterile inoculating loop to remove some of this film and prepare a streak plate on N-free agar. The streak plate will be incubated for a further 7 days at 30oC. 5. Examine wet mounts from your first streak plate culture using the phase contrast microscope. Prepare Gram stains of the film and look for large Gram negative cells that may be bacillus or coccoid in shape. They may occur singly, or in arrowhead-shaped pairs. Record observations in your lab book. 6. After the incubation period is complete, examine your streak plate. Look for large, translucent, mucoid colonies. Prepare a wet mount from an isolated colony and view it using a phase-contrast microscope. Prepare another streak plate using cells from the same colony. This plate will be incubated again, and observations will be made later in the term. Additionally, this culture will be used in Part B of this exercise. Thought Questions: 1. Define enrichment. What aspect(s) of the medium used in this exercise made it an enrichment medium? Why did we use the same medium for plating after freeliving nitrogen fixers were isolated? What term would we use to describe the medium in this case? 2. Why did we sample the film on top of the culture, rather than the sediment on the bottom of the flask? 3. When you viewed your Gram stains, you may have observed cells on your slides that didn’t appear to be Azotobacter. Why might these other genera be present? 16 PART B: IDENTIFICATION OF MICROORGANISMS USING PCR OF 16s rDNA The DNA from microbes can be isolated and may be studied via construction of BAC (Bacterial Artificial Chromosome) libraries (for an example, see Rondon, et al., 2000). More simply, an appreciation of diversity may be obtained by using universal primers for PCR amplification of rDNA genes from the Bacterial domain on a preparation of total DNA from an environmental sample. The resulting pool of nucleotide fragments may then be cloned, unique clones sequenced, and the resulting sequences analyzed in order to characterize and potentially identify the microbes present. In Part B of this exercise you will perform PCR using primers specific for prokaryotic 16s rDNA to isolate ribosomal DNA from putative Azotobacter cultures and then visualise this DNA using Agarose Gel Electrophoresis. In addition, DNA from successful PCRs will be sent for sequencing and you will then be using online tools to perform sequence analysis to confirm the identity of your cultures. PCR of Soil Bacteria Two different primer sets will be employed. Each group will only be using one set on their particular culture. Note that the primer designations refer to location of primer binding site on the 16s rDNA molecule. For preparation of your reaction mixtures: Benches 1, 3, and 5: Working with the people at your bench, each group will be setting up 3x reactions as outlined below: Primer Set #1 Template Source FP1/1492R Unknown Culture FP1/1492R E. coli K12 FP1/1492R No template Benches 2/4: Working with the people at your bench, each group will be setting up 3x reactions as outlined below: Primer Set #2 Template Source 27F/805R Unknown Culture 27F/805R E. coli K12 27F/805R No template 17 METHODS: Reagents: Taq (Invitrogen) 10x PCR buffer 50 mM MgCl2 Primers (*Y = C or T) FP1 (AGAGTTYGATYCTGGCT)*1 (10 pmol/L) RP1492 (TACGGYTACCTTGTTACGACT)*1 (10 pmol/L) 27F (AGAGTTTGATCCTGGCTCAG)2 (10 pmol/L) 805R (GACTACCAGGGTATCTAATCC)2 (10 pmol/L) dNTP mix (8 mM) Optima Water (Fisher Scientific) Cultures: Pure culture of organism isolated from soil Plate culture of E. coli Equipment: Thermocyclers (BioRad) Micropipettors and sterile tips Parafilm Ice buckets and ice Sterile 0.5 mL tubes Sterile 0.2 mL PCR tubes Biohazard bags Permanent markers Note: Use aseptic technique throughout. Keep your tubes on ice at all times! 1. Obtain three 0.2 mL PCR tubes from the sterile container at the side of the lab. Decide on appropriate codes for labeling the tubes (keeping in mind that other groups are carrying out the same reactions). Label the tubes on the tops and on the sides using permanent marker. Place the tubes on ice. 2. Obtain a 0.5 mL tube for your Master Mix. Keep this tube on ice. Use the information outlined in Table 1 to set up your Master Mix. This mix contains everything required in order for DNA replication to occur. Generally, Master Mixes contain enough volume to set up the number of reactions + 1. In your case, you will be preparing enough mix for 4 reactions. Work carefully. 18 Table 1. Components, starting concentrations and volumes for set-up of PCRs. Component and Starting Final Master Mix vol. Concentration Concentration (enough for 4 reactions) (L) Optima-Water 10x PCR buffer 50 mM MgCl2 dNTP mix (8 mM of all 4) Primer 1 (from set assigned to your bench) Primer 2 (from set assigned to your bench) Taq DNA polymerase (5 U/L) Template DNA 1x 1.5 mM 40 nmoles (0.8 mM of all 4) 0.4 M 128 20 8 20 8 0.4 M 8 5 U (units) 4 Leave Template out of Master Mix! Note: One primer set per reaction mixture! 3. While the Master Mix is being set up, other group members should be setting up template preparations. Obtain a small square of parafilm. For each bacterial culture (soil bacteria and E. coli – what is the role of E. coli?), use a micropipettor with a sterile tip to pipette 20 L of sterile Optima-water (Fisher Scientific) onto the parafilm. 4. Take a 10 – 100 L micropipettor and put on a sterile tip. Touch the tip to a single colony from your soil bacterial culture plate. Pipette up and down into the Optima water on the parafilm. 1 L of this mixture will be used as your template source. 5. Mix E. coli in the same fashion with your second drop of Optima water on the parafilm. Again, 1 L of this mixture will be used as template in your second reaction. 6. For your third reaction, you will be leaving out template and replacing it with an equal volume of sterile Optima water. What is the purpose of this reaction? 7. After preparation of Master Mix, add the appropriate volume of template or sterile water (1 L) to each tube, then check with the Instructor to see where everyone else is at. When all of the groups are at the same stage, add the appropriate volume of Master Mix (49 L ) to each tube. Keep your tubes on ice until in the PCR machine. GENTLY tap tubes to mix. When everyone is ready, the instructor will then show you how to operate the thermocycler. 19 The parameters you are using for the PCR are: 12 minutes at 95oC (used not only in initial DNA denaturation, but also to lyse the bacterial cells) 30 cycles of: 1 minute at 94oC 45 seconds at 55oC 90 seconds at 72oC A final elongation of: 20 minutes at 72oC The samples will be stored at -20oC upon completion. Thought Questions: 1. What are the purposes of the primers in PCR? 2. What happens at each temperature? 3. How is annealing temperature determined? 4. If you left out the forward primer, would you expect to see a band resulting on the gel? If you did, explain what this would mean. Suggested Background Reading: Amann et. al., 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59 (1): 143-169. Cole, J. R., Chai, B., Farris, R. J., Wang, Q., Kulam, S. A., McGarrell, D. M., Bandela, A. M., Cardenas, E., Garrity, G. M., and Tiedje, J. M. 2007. The ribosomal database project (RDPII): introducing myRDP space and quality controlled public data. Nuc. A. Res. 35: D169-D172. DeLong and Pace, 2001. Environmental diversity of bacteria and archaea. Syst. Biol. 50(4): 470-478. Gabor, E. M., deVries, E. J., and Janssen, D. B. 2003. Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS. 44(2): 153-163. Pace, 1997. A molecular view of microbial diversity and the biosphere. Science. 276: 734740. Whitford, M. F., Forster, R. J., Beard, C. E., Gong, J., and Teather, R. M. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe. 4: 153-163. Woese, C. R., Kandler, O., and Wheelis, M. L., 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA. 87: 4576-4579. 20 Agarose Gel Electrophoresis METHODS Reagents: 1x TBE buffer 0.8% agarose gels (1 per 2 benches) 10x loading dye 2-log NEB ladder premixed with loading dye Ethidium bromide bath PCR samples from last lab Equipment Power supplies (1 per 2 benches) Micropipettors Sterile tips Parafilm Transilluminator/camera Biohazard bags Gloves Note: Two groups will load their samples (6 tubes total) onto one gel. We will be using 0.8% agarose prepared in 1x TBE. 1. 2. Obtain and completely thaw your PCR samples. Using a micropipettor, 'dot' out 1 L aliquots of 10x loading dye in a line on a thin strip of parafilm. Remove a 7.5 L aliquot of your first sample, mix gently with the loading dye on the parafilm, and proceed with loading. Aim for approximately 1-2x final concentration of loading dye per sample loaded (and recognise that this is NOT exact). Loading Dye – 1) increases the density of the sample ensuring that it drops evenly into the well; 2) adds colour to the sample to simplify loading; and 3) contains dyes that in an electric field move toward the anode at predictable rates. In this laboratory, we are making use of mixtures containing xylene cyanol FF. This dye migrates in 0.5x TBE at approximately the same rate as linear DNA of 4000 bp in size. Often, bromophenol blue is used in conjunction with xylene cyanol, or separately. Bromophenol blue migrates at approximately the same rate as linear DNA of 300 bp in size in 0.5x TBE (2.2 fold faster than xylene cyanol FF, independent of agarose concentration). 3. 4. Load the remainder of the samples in the same manner, leaving at least one well empty (to be used for a DNA ladder). Be sure to RECORD the order in which the samples were loaded. Load 10 L of the ladder. 21 One type of size standard is produced by ligating a monomer DNA fragment of known size into a ladder of polymeric forms. The 2-log DNA ladder from New England Biolabs consists of a mixture of a number of proprietary plasmids digested to completion with different restriction enzymes. Ladders tend to be purchased as commercial preparations. For an example please see: http://www.neb.com/nebecomm/products/productn3200.asp 5. 6. Turn on the power supply and set the voltage to 100 V. Place the lid on the gel and start the run. The gel will run for 30 minutes, then shut off automatically. After the run is complete, turn off the power. Designate one group member to put on gloves, scoop up the gel, and gently slide the gel into the ethidium bromide bath. Caution: Ethidium bromide is a mutagen and a suspected carcinogen. At very dilute concentrations and with responsible handling, this risk is minimised. 7. Stain the gel with gentle shaking for approximately 10 minutes. One group member again should put on gloves, and transfer the gel to the gel documentation system. View using the UV transilluminator. Photographs will be taken. Please ensure that you bring a USB memory stick so that you can obtain the photograph of your gel (these will NOT be emailed out). Caution: Ultraviolet light is damaging to naked eyes and exposed skin. Always view through filter or safety glasses that absorb harmful wavelengths. 8. Based on gel results and quantification of your DNA, a selection of samples will be sent off for sequencing. In order to facilitate this, use a piece of tape to completely label your PCR products ensuring that the label corresponds with that from the gel. Thought Questions 1. Evaluate your gel results with respect to: expected fragment sizes and reasoning, and control results. Do we have evidence to suggest that we were successful in amplifying 16s rDNA? Explain your reasoning. 2. What are some of the advantages and disadvantages of molecular techniques for identification of bacteria? Compare and contrast with conventional culturing techniques. 3. After examining your sequence data, can you conclude that you isolated N-fixing organisms? Explain your reasoning. 4. Can you identify your unknown organisms to genus? To species? Why or why not? 5. What was the purpose of the control? How does it help you to make conclusions about your unknown isolates? 22 EXERCISE 4 SOIL AND COMPOST MICROBIAL ECOLOGY Soil Bacteria The microflora of the soil exist as a complex food chain that brings about the release of nutrients from dead plant material on the surface. The surface layer of newly fallen plant material is called the litter and chemically it is composed of insoluble materials such as cellulose, hemicellulose and lignin. Only a few organisms, usually fungi, are able to utilize these high molecular weight compounds since carbohydrates, amino acids, vitamins and other growth factors are lacking. However, once microorganisms do begin to decompose the litter, the chemical structure of the litter is modified and the organisms produce end products that are released into the environment and become available for use by others. Death of these organisms also provides new small molecular weight compounds that may then be utilized. Bacteria are able to utilize the end products of fungal metabolism. Nematodes and protozoa feed on the bacteria and mites and animals live on the nematodes and protozoa. In this way nutrients are recycled. Compost Bacteria Composting is a microbial process whereby plant matter including lignin is partially converted to humus, therefore supplementing the organic content of soil. The process is initiated by mesophilic heterotrophs and initially, is characterized by a temperature increase up to 55 – 60oC for a few days where thermophiles such as Bacillus stearothermophilus and Thermomonospora are active. The temperature then decreases, followed by several months of curing at mesophilic temperatures, where again, mesophiles predominate. Composting is not exclusively carried out by bacteria; fungi such as Aspergillus fumigatus, and Geotrichum candidum, are also involved. EXPERIMENTAL OBJECTIVES In this experiment, you will prepare serial dilutions of compost and of soil samples and plate out the appropriate dilutions. After incubation, you will determine the number of bacteria isolated in your two samples, and assess the microorganisms for their ability to utilise carboxymethylcellulose, casein, starch and xylan. You will choose one organism from either soil or compost, use biochemical tests to identify the microorganism you have chosen, and use the class results to compare and contrast microbial diversity in soil and in compost. Out of your group of four, one pair will prepare enumeration plates for soil, while the other pair will prepare enumeration plates for compost. 23 A. ENUMERATION OF BACTERIA IN SOIL AND COMPOST Period 1: 1. Weigh 1 g of soil or compost provided and add to 100 mL of sterile distilled water. This is dilution #1 (1:100). Shake the suspension for 5 minutes. For those pairs working with soil, please follow the instructions outlined in steps 2-5; those pairs working with compost, please follow steps 6-9. 2. Use the sterile 9 mL distilled water blanks provided to create serial dilutions of your soil. Please ensure that you vortex your samples well prior to making each new dilution. You will require 10-4 and 10-5 dilutions of soil. 3. Add 1 mL of the 10 -4 dilution to each of two sterile, labelled Petri dishes and 1 mL of the 10 -5 dilution to two labelled Petri dishes. 4. Obtain a 250 mL bottle of molten peptone yeast extract agar containing actidione from the water bath. (Actidione or cyclohexamide is an antibiotic that inhibits fungal growth). 5. Add approximately 20 mL of medium to the plates prepared in Step 3. Swirl carefully to mix the inoculum evenly with the medium. Label the bottle of molten agar with your group name and replace immediately in the waterbath. 6. Use the sterile 9 mL distilled water blanks to create serial dilutions of your compost. Please ensure that you vortex your samples well prior to making each new dilution. You will require 10-4, 10-5, and 10-6 dilutions of your compost. 7. Add 1 mL of the 10 -4 dilution to each of two sterile Petri dishes, 1 mL of the 10 -5 dilution to two sterile Petri dishes, and 1 mL of the 10-6 dilution to two sterile Petri dishes. 8. Obtain your labelled 250 mL bottle of molten peptone yeast extract agar containing actidione from the water bath. (Actidione or cyclohexamide is an antibiotic that inhibits fungal growth). 9. Add approximately 20 mL of medium to the plates prepared in Step 7. Swirl carefully to mix the inoculum evenly with the medium. Both pairs should make note of step 10: 10. The plates will be incubated for 24 hours at 30oC and refrigerated until the next lab session Period 2: Results Examine both sets of plates carefully and select the plates where the bacterial count ranges between 30 and 300 colonies. Record the number of colonies on the plates in number of bacteria per g of soil and of compost. 24 B. ISOLATION AND CHARACTERIZATION OF BACTERIA FROM SOIL OR COMPOST Each class will identify organisms from either soil or compost bacteria plates. Your instructor will indicate what plates to use. Work individually to complete Part B of this exercise. Period 2: 1. Choose a morphologically distinct colony from one of your plates (but approved by your instructor), and prepare a streak plate for single colonies on peptone yeast extract agar (Appendix 4). 2. The plates will be incubated for 24 hours at 30oC and then stored at 4oC. Period 3: 1. During your next lab period, prepare a Gram stain of the pure culture and record the cell morphology. Record the colony morphology of the culture (Appendix 6). 2. In addition, prepare new streak plates on PYE and NA and a broth culture using TSB from the colony you used to prepare your Gram stain. These cultures will be incubated at 30oC; PYE plates for 24 hours, and NA and TSB for 48 hours. Period 4: 1. Use the NA plate to prepare an endospore stain (Appendix 3). Use the broth culture and the phase contrast microscopes available at the back of the lab to look for the presence of capsules. 2. Subculture an isolated colony from your PYE plate into TSB – this culture will be incubated overnight at 30oC. You will use this culture as your inoculant for biochemical tests in subsequent periods. 3. Use the dichotomous key provided to develop a detailed outline in your lab book of the series of steps you plan to take to identify your unknown (this may require some out of lab time to complete). The outline should include tests to carry out as well as dates when you intend to do these tests. Your outline must be handed in and approved before you will be allowed to proceed. 25 The following media and reagents will be available for you to utilize as you attempt to identify your unknown: nutrient agar endospore stain reagents hydrogen peroxide (catalase test) oxidase reagent (oxidase test) IMViC reagents indole broths MRVP broths citrate slants urea slants litmus milk broths mannitol broth (with phenol red indicator) H & L medium containing glucose H & L medium containing lactose sucrose agar motility medium (with TTC) 26 1. Gram stain Gram positive ....................................................................................................... 2 Gram negative......................................................................................................16 2. Cell morphology bacillus or spirillum……......................................................................................... 3 coccus….............................................................................................................. 12 ovoid…................................................................................................. Azotobacter 3. Endospore stain positive………………………..................................................................................................4 negative…………………………............................................................................................. 6 4. 1True endospores……………………………............................................................................ 5 spore types……………………………....................................................................... 6 2 Special 5. Aerobe or facultative anaerobe………………………….............................................. Bacillus Obligate anaerobe........................................................................................ Clostridium 6. Bacillus cells may be branched, no true mycelium……………………………......................... 7 Bacilli form true mycelium……………………………............................................................ 10 7. Cells pleomorphic depending on age of culture…………………………................................ 8 Cells bacillus-shaped only. Club-shaped swelling may be present in young cultures…………………………....................................................................... 9 8. Cells pleomorphic becoming coccoid with age. Gram reaction of bacilli and cocci usually positive……………………...….................................... Nocardia Gram-positive coccoid cells in older cultures. Coccoid cells germinate to produce bacillus-shaped cells. Bacilli may be Gram negative with Gram positive granules…………………………............Arthrobacter 9. Catalase positive…………………………...................................................... Corynebacterium Catalase negative……………………………......................................................... Lactobacillus 10. Conidia or sporangia formed within one week…………………………............................... 11 No conidia formed, anaerobic or microaerophilic………………….................. Actinomyces 1 2 Endospores are seen within the vegetative cells on an endospore stain after growing on sporulation agar for 48 hours. Rod shaped cells break up into coccoid shapes or conidia after growing on an agar plate for several days. 27 11. Chains of conidia formed; colony may produce brown water soluble pigment and have an “earthy” smell……………....................... Streptomyces Single conidia only……………………………........... Micromonospora or Thermoactinomyces 12. Cells arranged singly, or in chains or clusters………………….………….............................. 13 Cells in cubical packets……………………………….........................................................…… 15 13. Catalase negative………………………………....................................................................... 14 Catalase positive………………………………........................................................................ 15 14. Large, mucoid colonies on sucrose agar; microaerophilic or facultative anaerobic; capsule present………………………....................... Leuconostoc Small, round (1-2 mm) colonies on sucrose agar, no capsule…………….... Streptococcus 15. Glucose fermented …………………………..................................................... Staphylococcus Glucose not fermented………………………............…........................................ Micrococcus 16. bacillus shaped…………………………….............................................................................. 17 coccus shaped…………………………....................................................................... Neisseria ovoid…………………………................................................................................ Azotobacter 17. Red or purple pigmented colonies on agar plate………………………............................... 18 No red or purple pigmented colonies; not associated with root formations in plants…………………………................................................................. 19 No red or purple pigmented colonies; associated with root formations in plants…………………………................................................................. 33 18. Acid from mannitol, pigment soluble in acetone:alcohol…………..………………..... Serratia No acid from mannitol…………………………............................................ Chromobacterium or Rhodopseudomonas or Rhodospirillum 19. Organisms produce a green, blue, brown or yellow water-soluble pigment which diffuses into the medium. Glucose respired; oxidase positive; aerobic; motile.......................................................................................... Pseudomonas No water-soluble pigment produced………………………................................................. 20 20. Curved or bent bacilli on Gram stain……………………………............................................ 21 Straight bacilli……………………………............................................................................... 22 21. Bent bacilli, methyl red (-), Voges Praskauer (+), catalase (+)…………….…………..... Vibrio Spiral bacilli, no growth in peptone water (indole broth) without cellulose strip………………………….................................................... Spirillum 28 22. Glucose not utilised……………………………...................................................................... 23 Glucose utilised facultatively……………………………........................................................ 25 Glucose utilised aerobically…………………………….......................................................... 28 23. Yellow pigmented colony…………………………........................................... Flavobacterium Non-pigmented colony………………………….................................................................... 24 24. Litmus milk alkaline, oxidase positive, aerobic, motile….............................. Alcaligenes Litmus milk alkaline, oxidase negative, non-motile…............................... Acinetobacter 25. Lactose fermentation produces acid…………………………............................................... 26 No acid from lactose……………………………..................................................................... 29 26. Methyl red (+), no growth on citrate, fecal odor on BHI……………………………. Escherichia Methyl red (-), growth on citrate…………………………..................................................... 27 27. Non-motile……………………………......................................................................... Klebsiella Motile…………………………............................................................................. Enterobacter 28. Yellow pigmented colonies…………………………..........................................Xanothomonas Non-pigmented colonies………………………..................................................... Acetobacter 29. Urease (+)……………………………...................................................................................... 30 Urease (-)……………………………...................................................................................... 31 30. Motile……………………………............................................................................................ 31 Non-motile……………………………........................................................................... Shigella 31. Indole (+), swarming growth on BHI agar…………………………................................ Proteus Indole (-), no swarming growth…………………………....................................................... 32 32. Litmus milk acid……………………………............................................................... Salmonella Litmus milk acid and peptonised…………………………....................................... Aeromonas 33. Citrate positive………………………….................................................................... Rhizobium Citrate negative……………………................................................................ Agrobacterium 29 Periods 5-8: Once your outline has been approved, carry out your tests using materials available in your kits or on the side bench. You will have 4 lab periods and possibly some open lab time to complete this part of the exercise. Record all of your results in your lab book. Include diagrams of all staining results, as well as descriptions of cell and colony morphology and tables of biochemical test results. Include the results from Part C below. Use reference material to identify your organism to species (note, identify all possible species). C. INVESTIGATION OF CATABOLIC ABILITY OF SOIL AND COMPOST BACTERIA This exercise will be completed concurrently with exercise B, above. Methods Each bench will require: 2 casein agar plates 2 NA plates containing carboxymethylcellulose 2 NA plates containing starch 2 NA plates containing xylan Each individual will require: 1 casein agar plate 1 NA plate containing carboxymethylcellulose 1 NA plate containing starch 1 NA plate containing xylan 4 replica-plating templates per bench sterile toothpicks – 1 beaker per bench waste beakers for used toothpicks Enumeration plates (of soil and of compost bacteria) Period 5: Each group of 4 is responsible for replica-plating 20 random colonies from plates of soil or compost bacteria onto plates containing one of the 4 substrates of interest (carboxymethylcellulose, starch, casein or xylan). Note that plates containing CMC, starch or xylan all contain these substrates added to a nutrient agar base whereas plates containing casein are composed of skim milk and agar only. 1. Using a sterile toothpick for each new colony, carefully scrape a well-defined colony from one of your soil or compost bacteria enumeration plates. 2. Stroke the toothpick across square number 1 on each of the three different labeled plates (CMC, starch or xylan). For plates containing casein as the substrate, you will need to estimate placement of the colonies as you will not be able to see the template through the plate. Place the used toothpick into the beaker provided. 3. Select a fresh toothpick and repeat steps 1-2 for 40 different colonies of bacteria. 30 Work individually to determine the catabolic ability of your unknown: 4. 5. Obtain 4 plates, each containing a different substrate. Label with your name and with the name of the substrate. Use an inoculating loop to streak out your unknown (from your PYE plate of pure culture) onto each of the 4 plates (you want to streak for single colonies). Invert all of the plates. These plates will be incubated at 30 oC for 48 hours. Period 6: Evaluation of Catabolic Ability: 1. For those plates containing xylan or carboxymethylcellulose as substrates, flood the plate with a 0.1% (w/v) aqueous solution of Congo Red. Incubate for 5-10 minutes, then pour off the excess solution into a waste beaker (not down the drain!), and flood the plate with 1M NaCl to destain. Swirl the plate and let stand. Over the next 30 minutes, perform this destaining step 2 more times. Be generous with the NaCl. Cellulase- or xylanase-producing colonies will be surrounded by yellow haloes visible against the red or orange background. 2. For those plates containing starch as a substrate, flood the plate with a 0.13% iodine/0.3% potassium iodine solution. Swirl to cover the surface of the plate, then discard immediately. Destain by flooding the plate with 1 M NaCl and allowing the plates to stand. Amylase positive colonies should be surrounded by a zone of clearing. 3. For those plates containing casein as a substrate, examine the plate closely. Caseolytic positive isolates should be surrounded by zones of clearing. Thought Questions: 1. For enumeration, why do you only count plates having between 30 and 300 colonies? 2. Why do you incubate soil and compost bacteria at 28-30oC? 3. Provide one specific example of a differential medium used in the current exercise. 4. What is the differential component in the medium in your answer in (a)? 5. How is the medium differential? 6. Is this medium type selective also? Why or why not? How would you make this medium selective for the carbon source in question? 7. In addition to manipulating nutrients, how else could you make culture conditions selective? Provide a specific example. 31 References: Atlas, R.M. and Richard Bartha. 1998 Microbial Ecology: Fundamentals and Applications, Fourth Edition. Benjamin/Cummings Publishing Company, Inc. 640 pp. Poulsen, O. M. and Petersen, L. W. 1989. Electrophoretic and enzymatic studies on the crude extracellular enzyme system of the cellulolytic bacterium Cellulomonas sp. ATCC21399. Biotechnol. Bioeng. 34: 59-64. Ross, H. 1993. Cellular, Molecular and Microbial Biology 343 Laboratory Manual. The University of Calgary press, Calgary AB. Teather, R. M. and Wood, P. J. 1982. Use of Congo red polysaccharide interactions in the enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43: 777-780. 32 EXERCISE 5 BACTERIAL REPRODUCTION Most bacteria reproduce by an asexual process called binary fission. In this process a single mother cell produces two identical daughter cells. Cell growth is often equated with increase in cell number due to the difficulty in measuring changes in cell size. Under ideal conditions populations of bacterial cells grow exponentially as cell number doubles at a regular interval or generation time (td). In the laboratory, pure cultures are routinely grown as batch cultures in test tubes and Erlenmeyer flasks. A batch culture is prepared by inoculating a fixed amount of liquid medium with the bacteria. The resulting culture is then incubated for an appropriate period of time with no further addition of microorganisms or growth substrates. Cell growth in batch cultures can be divided into four phases. Initially the culture is in a lag phase where cells are preparing to reproduce. During this time cells are adjusting their metabolism to prepare for a new cycle of growth. There is an increase in cell size without increasing numbers. As cells begin to divide and their growth approaches the maximal rate for the particular set of incubation conditions established, the culture enters the exponential growth phase (log phase). One cell gives rise to two, two cells give rise to four, and so on. In this phase, cells are growing and dividing at the maximum growth rate possible for the medium and incubation conditions. Growth rate is determined by a number of factors, including available nutrients, temperature, pH, oxygen and other physical parameters as well as genetic determinants. As nutrients become limiting or waste products accumulate, the growth rate once again slows and the culture enters the stationary phase. During this phase, there is no further net increase in cell number, as growth rate equals the rate of cell death. The final phase of a batch culture is the death phase. During this phase, there is an exponential decline in viable cell numbers. This decline may be reversed if environmental parameters are modified by the addition of nutrients, for example. The rate of growth of bacterial cells is usually monitored by measuring the increase in cell number. Bacterial cell numbers may be enumerated by a number of methods. Direct count methods enumerate all cells whether they are viable or not. The most common direct count method uses a microscope and a specialized counting chamber (e.g., PetroffHauser chamber) to count the number of cells in a known volume of culture. Automated systems such as Coulter counters may also be used to determine cell number. In contrast, indirect count methods require the growth of cells in culture in order to enumerate cell numbers. The most common method for enumerating living cells is the viable plate count. Serial dilutions of a cell suspension are prepared and spread on to the surface of a solid agar medium (spread plate) or incorporated into molten agar that is then poured into sterile petri dishes (pour plate). Following a suitable incubation time, the number of colonies growing on and in the inoculated agar are counted and used to 33 determine the number of viable cells in the original suspension. This method makes the assumption that each colony arose from a single viable cell or colony forming unit (CFU). Turbidimetric methods can be used to rapidly assess biomass (e.g., cell numbers). The amount of light passing through a cell suspension can be determined with a spectrophotometer. The optical density (OD) is a measure of the amount of light passing through the suspension. A calibration curve can be generated using suspensions of known numbers of bacteria. Prelab preparation: Turn on the spectrophotometer and set to 600 nm at least 15 minutes prior to taking readings. METHODS Ethanol/dettol in bottles Test tube racks Spectrophotometer blank containing LB broth Spectrophotometer Overnight culture of E. coli K12 strain 6 Culture flasks of LB (100 mL volume); pre-warmed 2 Culture flasks of LB with 2% NaCl; pre-warmed 2 Culture flasks of LB at pH 5; pre-warmed Please work in pairs. At 20 minute intervals, monitor the growth of your E. coli culture by optical density following the procedures outlined below. A. Culture inoculation: 1. Each group of two will be assigned one culture flask and one treatment (see Table below). Please mark the flask with your treatment, bench number and lab number, and replicate (A or B; to be assigned by your instructor). Groups in laboratory section 3 will continue to sample from the flask corresponding to your bench. Data from all three lab sections will be pooled and posted on the Biology 3400 web site. Table 1: Growth parameters for bacterial reproduction exercise. E. coli strain K12 K12 DH5 K12 K12 Medium LB LB LB LB with 2.5% NaCl LB at pH 5 Growth Temperature 37oC 30oC 37oC 37oC 37oC 34 2. Aseptically remove 1 mL of culture from the overnight E. coli culture you were assigned, and add it to your culture flask. Swirl to mix, and record an initial optical density value as described in Part B below. B. Culture sampling: 3. Everyone in the laboratory will be sampling at the same time. Samples will be collected four times during your regularly scheduled lab period at 20 minute intervals. For labs 1 and 2, these correspond to: 9:35 am (“time zero”), 9:55am, 10:15 am, 10:35 am, and for labs 3 and 4: 11:00 am, 11:20 am, 11:40 am, and 12:00 pm. Your laboratory instructor will set a timer so that everyone is coordinated. Just prior to your readings, zero the spectrophotometer as outlined in Appendix 7. 4. At the times indicated, carefully remove your flask from the shaking incubator, remove 5 mL of your culture, and place it into a spectrophotometer tube. Replace your culture flask back into the incubator, and then read the absorbance of the sample you collected. Record the optical density (Absorbance) reading in your lab book and in the spreadsheet. 5. Your lab instructors will collect and distribute class data. Prepare graphs for each of the parameters tested, and use them to address the thought questions below. Thought Questions: 1. Use your graphs to calculate generation time of E. coli in each of the cultures. 2. What impact does increasing the concentration of NaCl have on growth of E. coli? 3. Do changes in the pH or temperature cause a change in the rate of growth of E. coli? Provide reasons for any changes observed. 4. Are there differences in the rate of growth of the two strains of E. coli examined? Why might this be? 5. Compare your values to that from the literature. Do the values differ? Why might this be? 35 EXERCISE 6 FERMENTATION A number of industrial processes make use of the end products of bacterial and fungal fermentations. For instance, in the presence of acid producing bacteria, and often the enzyme renninase, milk will form curds (solid) and whey (liquid). Once the solids are compressed, salted, and aged, the resulting product is cheese. Different cheeses are produced by varying the bacterial inoculum, varying the milk used, or even by introducing fungi such as certain species of Penicillium into the curds. Some Streptococcus species and some Lactobacillus species produce only lactic acid as a result of reduction of pyruvic acid. These organisms are responsible for the production of yogurt. Yogurt can be made from milk simply by inoculating with a starter culture of yogurt that contains live bacterial culture. Conversely, yeasts produce alcohol and CO2 rather than lactic acid as a result of the reduction of pyruvic acid. We are going to examine the latter two processes in this exercise. Part A: Yogurt Production MATERIALS Milk Skim milk powder Balance Erlenmeyer flasks – 500 mL Hot plates Thermometers Beakers – 250 mL Bacterial inoculant Probiotic capsules Water baths Micropipettors and tips pH meter phenolphthalein 0.1N NaOH Procedure – Period 1 1. Work as a bench group to heat approximately 250 mL of milk to 85 oC using the Erlenmeyer flask and hot plate provided. Hold the temperature at 85 oC for 2 minutes, and then carefully remove the flask from the hot plate, and let cool slowly on the bench to about 42-45oC. 2. Transfer 125 mL of heated milk into each of two beakers. Each pair at the bench will then be responsible for preparing one of the treatments listed in Table 8.1 below. Inoculant “A” is live yogurt culture. Add one rounded teaspoonful to 125 mL milk samples. Inoculant “B” is a commercial yogurt bacterial started – add 36 0.63 g to milk samples. Inoculant “C” is a probiotic capsule, which contains some of the same bacterial species as those found in the commercial starter. Empty the contents of one capsule into the cooled milk sample for treatment 11. 3. For treatments where skim milk is added, mix 6 g into the cooled milk prior to adding the bacterial inoculant. Add the inoculant, and gently stir to mix. Label your beakers and place them in the water bath appropriate for your treatments. Your instructor will remove beakers after approximately six hours of incubation, and cultures will be stored at 4oC until the next lab period. Table 6.1: Experimental set-up for yogurt cultures; A = yogurt, B = inoculant, C = probiotic. Treatment 1 2 3 4 5 6 7 8 9 10 11 12 Inoculant A A A B B B A B A B C none Skim Milk Powder no no no no no no yes yes no no no no Incubation Temperature 32oC 42oC 52oC 32oC 42oC 52oC 42oC 42oC 42oC-with shaking 42oC-with shaking 42oC 42oC Procedure – Period 2 1. Examine all yogurt cultures (not just the one you set up), and make observations regarding texture, consistency, smell and general appearance in your lab book. 2. Determine the pH of your cultures using the pH meter provided. 3. The amount of acid produced during the fermentation process can be calculated by titrating your culture with 0.1 N NaOH in the presence of phenylphthalein, a pH indicator. Measure 9 mL of culture and add to a small beaker, along with 4 drops of indicator. Mix well. Slowly add NaOH until a sustainable colour change is observed. Record the volume of NaOH required, and multiply by 0.1 to convert to % lactic acid. 4. Record your data in the table on the board and in your lab book. 5. Prepare a Gram stain of your culture and record observations in your lab book. How many different types of organisms do you see? 37 Part B: Alcohol Production from Yeast Prior to your lab period, grape juice, water and yeast cells were added to a sterile container. Over the next ten days, you will be responsible for sampling the fermenting juice at various time intervals. The primary fermentor is inoculated with a high cell density (~106 yeast cells/mL). The bulk of the must (grape juice medium) is rapidly depleted of oxygen by the yeast and remains anaerobic, despite the primary fermentor remaining open to the atmosphere. Yeast cells continue to reproduce by acquiring the needed energy and carbon through fermentation. The fermentation is an ethanolic fermentation because ethanol and CO2 are the fermentation endproducts. Growth of the yeast culture can be monitored by measuring optical density and enumerating CFU/mL (Recall Exercise 5 - Bacterial Reproduction). Ethanol concentration can be estimated indirectly by measuring the specific gravity of the wine must with a hydrometer. The specific gravity of the must decreases as the grape juice sugars are converted to ethanol and CO2. A "specific gravity to percent ethanol" conversion chart supplied with the hydrometer is then used to determine ethanol content of the must. Note: this exercise will require some out of lab participation. Please sign up for two time slots when at least half of your group members can attend. MATERIALS (in C741) Wine thief Spectrophotometer (warm up 15 minutes prior to reading OD values) Primary fermentor pH paper Micropipettors and sterile tips Sterile microfuge tubes Rack for microfuge tubes Filter sterile wine must for diluting samples Bunsen burner Hydrometer Sterilising solution Graduated cylinder Thermometer 30oC incubator YPD plates Spreaders and alcohol Spreadsheet for recording results 38 At each time point the following data must be collected by each group: Temperature OD600 pH Specific gravity Viable counts Procedure 1. Sterilize (by immersion in metabisulfite), the spoon, the wine thief, the sampling graduated cylinder, the hydrometer and the thermometer, and rinse each in water prior to touching the wine. Use the sterilized spoon to stir the mixture in the primary fermentor. 2. Use the sterilized wine thief to remove mixture from the primary fermentor into the plastic graduated cylinder. Add enough sample to bring the level up to the line marked. 3. Measure and record the sample temperature in the cylinder. 4. Measure and record specific gravity. Use the diagram posted to assist you. 5. Aliquot some mixture into the 125 mL Erlenmeyer flask provided – label with sampling time, and place in the tray in the deli cooler. An instructor will read and record the pH for you. 6. Use a sterile 5 mL pipette to remove 5 mL of sample from the cylinder and place into a sterile spec tube. 7. Remove the 50 mL Falcon tube of sterile wine must labeled with your time point from the deli fridge, and use it to make appropriate dilutions. 8. Use the blank provided to zero the machine. Read the optical density of your sample. If the OD600 exceeds 0.7, you will have to dilute the sample with the sterile must provided (start by creating a 1:1 dilution). Read the OD 600 of the diluted sample, then multiply by the dilution factor to obtain your corrected reading. Record the corrected reading on the sheet provided. 9. Viable counts: Prepare in duplicate. Refer to the posted table to determine the number of dilutions required. Prepare required dilution series – check dilution calculations with your lab instructor first. Label plates with time, name and dilutions. Spread plate (in duplicate) 100 µL of suggested dilutions on YPD agar. Invert plates and incubate at 30ºC – incubator is labeled. Plates will be removed and placed into the fridge. During lab, you will count and record values from plates having between 30 and 300 colonies. Tidy up work area!!– put spec tubes into labeled rack, discard tips, pipettes and microfuge tubes into Biohazard bag, put everything back where you found it. Pour out wine sample and resterilize cylinder for the next group. 39 Thought Questions (to be answered in your notebooks): 1. Describe the metabolic reactions that result in the production of yogurt. What microorganism(s) is/are required? How does each play a role? 2. Explain the effects of the following on the fermentation process that occurs during yogurt production: addition of skim milk powder, incubation temperature, shaking and type of inoculant used. 3. What is the purpose of the heat pre-treatment in the yogurt making procedure? 4. What was the purpose of incubating the treatment containing milk only? 5. Numerous data relating to alcohol fermentation were collected by the class over the sampling period, including measurements of pH, temperature, specific gravity, optical density, and CFU’s/mL of culture. Design and construct a series of figures to graphically represent the data that were collected. 6. Calculate the generation time of the yeast cells in the culture. 7. Name two factors that control the final ethanol concentration in a culture. 8. Although we stirred our culture each time before sampling, winemakers do not. Why would winemakers not stir the culture? 9. Why did the pH of the culture change as fermentation proceeded? 10. Why did the specific gravity of the culture change over time? 40 EXERCISE 7 VIROLOGY The objectives of this series of exercises are first to isolate coliphage from filtered raw and treated sewage obtained from the Lethbridge Wastewater Treatment Plant, to examine the plaque morphologies, and to prepare phage isolate from one particular plaque. Using this phage isolate, the phage titre will be determined, and the host specificity of the phage will be examined using several enteric bacterial strains. These exercises will demonstrate standard techniques in phage isolation and manipulation. (Please review the material on sewage treatment posted on the Biology 3400 web page). Prior to the laboratory, sewage samples were collected at the areas indicated on the schematic posted on the web page. Both samples were stored at 4 oC prior to filtering, for up to 1 week. On the morning of the lab, samples were filtered twice using 0.45 m filters. PART A - ISOLATION METHODS: For each bench: Luria Methylene Blue agar plates Overnight culture of Escherichia coli LE 392 Bottle of molten Luria agar overlay (at 60 oC) Sterile test tubes Test tube rack Micropipettor (100 L – 1000 L) Sterile tips Microbiology kits For the lab: Vortex mixer Water bath set to 60 oC Raw and treated sewage filtrate Test tube showing 4 mL mark Work in groups of 4. Note that sewage filtrate contains human pathogens. Work very carefully. Students who are clearly unprepared or are sloppy will be asked to leave the lab. 41 Period 1 - Procedure 1. Obtain a tube of culture of E. coli LE 392. 2. Obtain 5 Luria Methylene Blue agar plates, and 5 sterile test tubes. Label your 5 tubes according to Table 7.1. Table 7.1 Experimental set-up for isolation of coliphage from sewage. Tube # Contents (L) LE 392 Raw Sewage Filtrate Treated Sewage Filtrate 1 500 0 0 2 0 500 0 3 0 0 4 500 500 0 5 500 0 500 500 3. Pipette the appropriate amount of filtrate and/or cells into each of your labeled test tubes. Leave the tubes at room temperature on your bench to incubate for 20 minutes to allow the phage to adsorb to the cells. 4. While your cultures are incubating, label your Luria Methylene Blue plates according to Table 7.1. Mark the level of 4 mL on each of your tubes using the marked test tube on the side bench as a guide. 5. Starting with Tube 1, aseptically pour molten agar into the tube up to the level of 4 mL. Vortex to mix, then immediately pour the contents over the surface of the appropriately labeled plate. Swirl the plate gently to ensure that the entire surface is covered with the agar. 6. Repeat step 5 for the remaining tubes and plates. 7. After 10 minutes, the overlay should be set. Invert your plates and place them on a tray on the side bench to be incubated. Plates will be incubated at 37 °C for 16 – 20 hours, then stored at 4 °C until the next laboratory period. The next laboratory: Work in groups of four. MATERIALS Pasteur pipettes Bulbs Gloves Chloroform (in the fume hood) Vortex mixer Phage dilution buffer 42 Plates from last lab 1 dissecting microscope per bench Microfuge tubes (sterile) 1 mL pipettes and propipettors Microfuge racks Labeled microfuge rack on the side bench for class tubes Period 2 - Procedure 1. Obtain your plates. Examine them carefully. Record the number of plaques present for both raw and treated filtrate. Is there any difference? 2. Make detailed observations of plaque morphology. Features to look for include size, shape, and turbidity (clear vs cloudy). Use the dissecting microscopes for your observations. 3. After making observations, obtain a microfuge tube and aseptically add 1 mL of phage dilution buffer to your tube. Label with your group designation. 4. Use a Pasteur pipette (with a rubber bulb attached) to remove a plaque (squeeze the bulb, insert pipette into the agar over a plaque, gently release bulb to remove a plug of agar containing the plaque). Release plaque into the prepared tube of phage dilution buffer. 5. Vortex vigorously to disperse the agar. 6. Move to the fume hood and use a Pasteur pipette to add a drop of chloroform to your tube. Vortex the mixture once again. What does the chloroform do? 7. Place your tubes in the rack on the side bench. The tubes will be stored at 4 oC allowing the phage to elute from the agar into the buffer. PART B – AMPLIFICATION OF PHAGE BY PLATE LYSIS METHODS Cultures/phage: Overnight culture of E. coli strain LE 392 Tube containing plaque isolated previous day Other supplies: Waterbaths set at 37 oC, 60 oC Test tube racks in the 37 oC waterbath 15 mL Falcon tubes Top agar (molten in 50 oC waterbath) Freshly prepared methylene blue LB plates at room temperature vortex mixers Biohazard disposal bags 43 Period 3 – Procedure For Plating your Phage: 1. Obtain two freshly poured methylene blue LB plates. Label appropriately. 2. Obtain two test tubes per bench. One is for phage plus cells while the other is for a control. Label appropriately. Aseptically, transfer 0.1 mL of E. coli LE 392 culture to one tube. 3. To your cells in the tube, add 1/10 of the volume of your buffer (0.01 mL) containing the plaque. 4. Set up a negative control containing E. coli only in the same fashion. 5. Place the tubes in the 37 oC waterbath and incubate here for 20 minutes. What is the purpose of this incubation step? 6. Ensure you have all supplies for plating close at hand. Aseptically, add 3 mL of top agar to each tube. 7. Working quickly, vortex briefly to mix phage, cells and top agar, then pour over the surface of the labelled plate corresponding to the mixture. Swirl the plate to evenly distribute the mixture over the entire surface. Do not swirl for more than a few seconds! 8. Leave the plates face-up to dry. 9. Transfer the plates also face-up to the labelled tray on the side bench. Plates will be incubated without inversion for 16-24 hours at 37 oC. For Harvesting Phage: Note: One member of each group will be required to come in make observations and add buffer to plates tomorrow (out of regular lab time) Additional Supplies Chloroform (in the fume hood) -* Caution: Chloroform is toxic* Falcon tubes (polypropylene) Shaker at 4 oC 5 mL pipettes and propipettors Phage dilution buffer Pasteur pipettes and bulbs 100 L and 1 mL micropipettors and sterile tips Biohazard bags Centrifuge at 4 oC Vortex mixer in fume hood Balance and beaker The day prior to lab: 1. View your plates and record observations for both the control and phage treatments. 2. Aseptically add 5 mL of phage dilution buffer to the surface of each plate containing phage and cells. 3. Place these plates on the shaker at 4 oC to shake gently overnight. 44 Period 4 - Procedure 1. Using a Pasteur pipette, transfer as much of the liquid as possible into a sterile Falcon tube. 2. Move to the fume hood and add 0.1 mL of chloroform to each tube. 3. Vortex briefly, then find another group at the same stage for balancing your tubes. 4. Balance your tubes to within 0.1 g as follows: place 1 tube in a beaker on the balance pan and zero the balance remove the first tube and place the second in the beaker. Note the mass. adjust the volume in the tubes with sterile phage dilution buffer such that the balance reads the same for both tubes. 5. 6. Place your tubes into the centrifuge in a balanced configuration; ie, the 2 tubes balanced against each other should be across from each other. Centrifuge at 4000 g for 10 minutes at 4 oC. PART C – HOST RANGE AND PHAGE TITRE METHODS Overnight cultures of: E. coli strains CSH121 and CSH125 and LE 392 Enterobacter Other supplies: Phage dilution buffer Micropipettors and sterile tips Autoclave waste disposal Luria Methylene Blue agar plates LB plates Bottle of molten Luria agar overlay (at 60 oC) Sterile test tubes Test tube indicating 4 mL mark Test tube rack Micropipettor (100 L – 1000 L) Sterile tips Microbiology kits For determining phage titre: 1. Prepare serial dilutions of your phage in dilution buffer (10 -2, 10-4, 10-6, 10-8) in microfuge tubes. Vortex each tube as you create each dilution. Ensure that you use fresh tips for each transfer. 2. In separate, labeled sterile test tubes, mix 500 L of each dilution (10o, 10-2, 10-4, 10-6, 10-8) with 500 L of host strain E. coli LE 392. Sit for 20 minutes of incubation time at room temperature. Mark the 4 mL mark on each test tube while mixtures are incubating. 3. Plate your mixtures as per Part A of this exercise. 45 4. The next day, count plaques and determine the titre of your phage. For determining host range: 1. Prepare spread plates on LB for each organism to be tested. (use the instructions found in Appendix 3, although this should be a review from previous courses). Label each plate clearly. Use 100 L of liquid culture to create a uniform lawn. 2. When lawns are dry, divide plates into four quadrants. Spot 20 L of undiluted phage, or 20 L of your 10-2, 10-4 or 10-6 dilution in each quadrant. Do not invert. Plates will be incubated at 37 oC overnight. Period 5 - Procedure 1. The next day, score as + or – for phage growth on each host. Thought Questions: 1. Based on the schematic found on Dr. Brent Selinger’s web site, what step(s) is/are most likely responsible for the difference in coliphage numbers between raw and treated sewage? 2. Have you isolated more than one type of phage? How might you be able to tell? 3. To what components of the bacterial cell do phage typically adhere? 46 APPENDIX 1 MICROSCOPY To view microscopic organisms, their magnification is essential. The microscope is the instrument used to magnify microscopic images. Its function and some aspects of design are similar to those of telescopes although the microscope is designed to visualize very small close objects while telescopes magnify distant objects. Please review Appendices 1 and 9. Magnification is achieved by the refraction of light travelling though lenses, transparent devices with curved surfaces. In general, the degree of refraction, and hence, magnification, is determined by the degree of curvature. However, rather than using a single, severely-curved biconvex lens such as that of Leeuwenhoek's simple microscopes, Hooke determined that image clarity was improved through the use of a compound microscope, involving two (or more) separate lenses. Properties of the Objective Lenses 1. Magnification Magnification is a measure of how big an object looks to your eye. The number of times that an object is magnified by the microscope is the product of the magnification of both the objective and ocular lenses. The magnification of the individual lenses is engraved on them. Your microscope is equipped with ocular lenses that magnify the specimen ten times (10X), and four objectives which magnify the specimen 4X, 10X, 40X, and 100X. Each lens system magnifies the object being viewed the same number of times in each dimension as the number engraved on the lens. When using a 10X objective, for instance, the specimen is magnified ten times in each dimension to give a primary or "aerial" image inside the body tube of the microscope. This image is then magnified an additional ten times by the ocular to give a virtual image that is 100 times larger than the object being viewed. 2. Resolution Resolution is a measure of how clearly details can be seen and is distinct from magnification. The resolving power of a lens system is its capacity for separating to the eye two points that are very close together. It is dependent upon the quality of the lens system and the wavelength of light employed in illumination. The white light (a combination of different wavelengths of visible light) used as the light source in the lab limits the resolving power of the 100X objective lens to about 0.25 µm. Objects smaller than 0.25 µm cannot be resolved even if magnification is increased. Spherical aberration (distortion caused by differential bending of light passing through different thicknesses of the lens center versus the margin) results from the air gap between the specimen and the objective lens. This problem can be eliminated by filling the air gap with immersion oil, 47 formulated to have a refractive index similar to the glass used for cover slips and the microscope's objective lens. Use of immersion oil with a 100X special oil immersion objective lens can increase resolution to about 0.18 µm. Resolving power can be increased further to 0.17 µm if only the shorter (violet) wavelengths of visible light are used as the light source. This is the limit of resolution of the light microscope. The resolving power of each objective lens is described by a number engraved on the objective called the numerical aperture. Numerical aperture (NA) is calculated from physical properties of the lens and the angles from which light enters and leaves. Examine the three objective lenses. The NA of the 10X objective lens is 0.25. Which objective lens is capable of the greatest resolving power? 3. Working Distance The working distance is measured as the distance between the lowest part of the objective lens and the top of the coverslip when the microscope is focused on a thin preparation. This distance is related to the individual properties of each objective. 4. Parfocal Objectives Most microscope objectives when firmly screwed in place are positioned so the microscope requires only fine adjustments for focusing when the magnification is changed. Objectives installed in this manner are said to be parfocal. 5. Depth of Focus The vertical distance of a specimen being viewed that remains in focus at any one time is called the depth of focus or depth of field. It is a different value for each of the objectives. As the microscope is focused up and down on a specimen, only a thin layer of the specimen is in focus at one time. To see details in a specimen that is thicker than the depth of focus of a particular objective you must continuously focus up and down. How to use a compound light microscope: Locate the ocular lens (eyepiece); there will be one if the microscope is monocular, or two if it is binocular. Then locate the objective lenses, the ones nearest the object to be studied. These two lenses (ocular and objective) are connected by the body tube of the microscope. The objective lenses (there will be two or more, the smallest being that with the least magnifying power, and the largest being that with the greatest magnifying power) are mounted on a revolving nosepiece above a flat stage on which the study specimen (slide) is placed. 48 Figure 1: The Compound Microscope Your microscope is equipped with a mechanical stage. This consists of a clip to hold the slide in place (the clip is spring-loaded; the Instructor will demonstrate how it works) and two knobs at the side of the microscope body to move the slide side-to-side, or forwardto-back. Note also the two micrometer scales on the mechanical stage, which allow you to note the coordinates of a particular object on the slide you are viewing. Place a slide on the stage and center it over the hole in the stage. Adjust the distance between the oculars to match your interpupillary distance (distance between your pupils). Revolve the nosepiece so that the lowest power objective lens (generally the 10x power lens) is in position. To focus the microscope, locate the coarse and fine adjustment knobs at the base of the microscope, and use the coarse adjustment to move the slide close to, but not touching, the objective lens. Look at the stage from the side as you do this. On most microscopes this involves raising the stage, but on some the lenses are lowered. Also, on most microscopes an automatic stop will prevent you from moving the stage closer than about one centimeter from the lens. Now, look through the ocular lenses, and move the slide away from the objective lens until the specimen becomes clear (is in focus). Finish focusing with the fine adjustment knob. Once you have focused with the low objective power lens, you may switch over to the next higher power lens with only fine focus adjustments (the microscope is said to be parfocal). 49 As you switch from one objective lens to another, you will notice that the working distance, the clearance between lens and stage, decreases with increasing lens power. This is illustrated in Figure 2 below. Figure 2: The working distance (above) and the field of view (below) change with magnification of objective lens. It should be obvious to you why, on high power objective lenses (40x or 100x), you must use only the fine focus knob to adjust focus; otherwise the risk of (damaging) contact between lens and slide becomes great. Also illustrated in Figure 2 is the diminishing field of view as objective lens power increases; this is due to a smaller and smaller aperture at the bottom of the lens through which light enters. This means that [a] things are harder to find on a slide when you are using high power since only a small fraction of the slide can be seen, and [b] less light enters your eye and everything in the field appears darker. As a consequence, you will learn to [a] switch back to a lower power objective lens when you want to "scan" around the slide, and [b] manipulate the amount of light coming into the lens so that you can see the objects clearly. The amount and concentration of light coming through the specimen and hence to your eye can be adjusted in several ways. First, of course, is the on/off light switch, generally located at the base of the microscope, and often associated with a rheostat to control light intensity. A condenser lens is mounted below the stage, and concentrates the light on to the specimen; it generally needs no adjustment of position. An iris diaphragm is located below the condenser lens. Find the lever which controls the diaphragm; it can be very useful in adjusting illumination and contrast. 50 Biology 3200 microscopes are binocular, containing two eyepieces. To correct for the slight difference in the focus of your two eyes, precisely fine focus a specimen using only your one eye which is at the non-focusing ocular (if your microscope contains two focusing oculars, either may be used to begin). Next, open the other eye and bring the image into focus for that eye using only the ocular focus. Since other students use these same microscopes during the semester, this exercise of binocular focusing should be performed at the onset of each microscope session. Finally, some useful hints and cautions: Never drag the microscope across the counter-top. Lift it with both hands by its arm, being careful not to tip it. Use lens paper to clean glass slides and lens surfaces before using your microscope. Water damages objective lenses; if water does contact a lens, wipe it off immediately. Also avoid getting water under the slide as it will stick to the stage. If you have used immersion oil, use lens paper dipped in 60 % ethanol to remove it from the 100x objective lens when you are finished. Always start the focusing procedure with low (10x) power lens. When attempting to locate an object on a slide, remember that the image you see is reversed; that is, as you move the slide toward you on the stage, the slide is apparently moving away from you as you view it through the lens. Some ocular lenses are equipped with pointers; they appear as a dark black line that will rotate if the lens is rotated in its tube. Phase Contrast Microscope The phase contrast microscope was developed by Frits Zernike, a Dutch mathematical physicist, in 1936. His discovery, for which he was awarded a Nobel Prize in 1953, led to the development of other types of microscopes (confocal and fluorescence microscopy). Phase contrast microscopes are based on the principle that cells differ in refractive index from their surroundings, meaning that light that passes through a cell differs in phase (the light is slowed as it passes through a cell) from light passing through its surroundings. The difference in phase is subtle, but can be enhanced, or amplified by a device called a phase ring. The result is a dark image on a light background. Phase contrast microscopy is a very powerful tool for observing living cells and their activities. Tips for use: Prepare a wet mount slide, place it on the stage, and find and focus on the cells using brightfield optics at a low-power magnification. Once the image is in focus, move to the next highest magnification. Switch to phase optics that match your objective lens (coding is present on the objective lenses) by rotating the disc underneath the stage. 51 The use of phase contrast requires much more illumination as much of the light is lost as it passes through the phase ring. Increase the amount of light hitting the specimen once you switch to phase. Switch to higher magnifications in the same way that you would for brightfield optics, but remember to rotate the annular stop corresponding to the objective lens into position each time. Care and Feeding of the Microscopes Checklist For Compound Microscopes Name:________________________________________ Class and section: ______________________________ Date:_________________________________________ Microscope #: _________________________________ Did you find the microscope in proper working order? Y or N If not, what was the problem? _____________________________________________ _____________________________________________ _____________________________________________ _____________________________________________ __ Slide removed from stage __ Slide, stage, and objectives are free of oil __ Mechanical stage is centered __ Stage placed at its lowest position __ 4x objective placed into working position __ Ocular micrometer replaced with regular ocular __ Binocular head secured in “start” position __ Rheostat turned to 0 and lamp is turned off __ Cord is wrapped tightly around arm and lamp __ Cord is secured with cord clip __ Dust cover is placed over scope 52 Checklist For Dissecting Scopes Name:________________________________________ Class and section: ______________________________ Date:_________________________________________ Microscope #: _________________________________ Did you find the microscope in proper working order? Y or N If not, what was the problem? _____________________________________________ _____________________________________________ _____________________________________________ _____________________________________________ __ Turn off transformer __ Unplug transformer and lamp __ Wrap cord tightly around transformer __ Place transformer on stage with binocular head well above the transformer __ Replace dust cover 53 APPENDIX 2 PREPARATION OF SCIENTIFIC DRAWINGS Use a sharpened pencil; never ink. The lead should be hard. Place drawing to one side, usually the left, leaving room for labels to the right. Try to draw with one continuous line and do not retrace your lines. Do not shade. Place label lines horizontally (use a ruler), with no crossed lines. Objects labelled should be singular unless label line branches to multiple objects. Label only what you see, not what you think should be seen. Below the figure you should add: The title of the diagram The magnification of the drawing (see below) The magnification of the diagram gives you the relationship between the size of your diagram and the actual size of the specimen. A diagram of a cell would be much larger than the actual cell, whereas a diagram of an elephant could be much smaller than the actual elephant. Magnification is defined as: size of drawing actual size of specimen Where: size of the drawing is measured with a ruler actual size of specimen is determined by one of the methods in Exercise 1. the number calculated has as many significant figures as the accuracy of your measurement (usually 2, if you measure in mm) 54 Example of a drawing: Figure 1. A chain of Bacillus subtilis cells stained with methylene blue (23 000x) Notice that in the figure, enough organisms are shown such that the arrangement can be seen. Drawing magnification is calculated based on length or width, not both of only one of the organisms (not the whole chain). Figures are given numbers - Figure 1, Figure 2, etc. As much detail as possible is provided in the title (eg Gram reaction seen, type of stain used, type of organism etc.). 55 APPENDIX 3 STAINING Bacteria cells are very difficult to observe using compound light microscopes because the cells appear transparent in the aqueous medium in which they are suspended. Staining the cells prior to observation increases the contrast between the cell and the medium, which allows for the visualization of cell structures. However, the application of stains usually leads to cell death. Phase contrast microscopes enhance the contrast between cells and their environment without the use of stains, meaning that living cells and their activities can be observed. We will use both approaches to study the morphology of microorganisms in this exercise. Staining In general, prior to any staining procedure, fixation occurs. Fixation performs two functions: (i) immobilizes (kills) the bacteria; and (ii) affixes them to the slide. Any procedure that results in the staining of whole cells or cell parts is referred to as positive staining. Most positive stains used involve basic dyes where basic means that they owe their coloured properties to a cation (positively charged molecule). When all that is required is a general bacterial stain to show morphology, basic stains such as methylene blue or carbol fuchsin result in the staining of the entire bacterial cell. Differential stains are used to distinguish bacteria based on certain properties such as cell wall structure. Differential stains are useful for bacterial identification, contributing to information based on bacterial size, shape, and association. Differential staining relies on biochemical or structural differences between the groups that result in different affinities by various chromophores. Gram staining behaviour relies on differences in cell wall structure and biochemical composition. Some bacteria when treated with para-rosaniline dyes and iodine retain the stain when subsequently treated with a decolourising agent such as alcohol or acetone. Other bacteria lose the stain. Based on this property, a contemporary of Pasteur, Hans Christian Gram, developed a rapid and extremely useful differential stain, which subsequently bears his name - the Gram stain used to distinguish two types of bacteria, Gram positive and Gram negative. Gram negative forms, which are those that lose the stain on decolourization, can be made visible by using a suitable counterstain. The strength of the Gram stain rests on its relatively unambiguous separation of bacterial types into two groups. However, variables such as culture condition, age or environmental condition, can influence Gram staining of some bacteria. 56 The bacterial cell wall is very important for many aspects of bacterial function and hence, the Gram stain also provides valuable information about the physiological, medicinal and even ecological aspects of the bacteria. Negative staining is used to characterize external structures, like capsules, that are associated with living bacterial cells. Negative stains make use of acidic dyes where acidic means that they owe their coloured properties to an anion (negatively charged molecule), so they are repelled by the negatively charged cell wall. Hence, the cell is transparent and its surroundings are coloured. Negative staining is useful for determining cell dimensions and visualizing capsules, as heat fixation shrinks both cells and capsules. Poly--hydroxybutyric Acid (PHB) Staining PHB granules are common inclusion bodies in bacteria. Monomers of hydroxybutyric acid are connected by ester linkages forming long polymers which aggregate into granules. As these granules have an affinity for fat-soluble dyes such as Sudan black, they can be stained and then identified with the light microscope. These granules are storage depots for carbon and energy. Endospore Staining Certain bacteria may produce endospores under unfavourable environmental conditions. Endospores are mainly found in Gram-positive organisms, including the Gram-positive Clostridium and Bacillus, in the Gram-positive cocci Sporosarcina, and in some of the filamentous Gram-positive Monosporaceae family. It has also been discovered that Coxiella burnetii, a small rod found in raw milk that has a variable Gram stain reaction, but a typical Gram-negative cell wall has a sporogenic cycle. When conditions become more favourable, the endospores will germinate and the bacteria will return to the actively growing and dividing form. Endospores are highly resistant to heat, chemical disinfectants and to desiccation and therefore allow the bacterial endospore to survive much more rigorous conditions than the vegetative cells. Endospore resistance is due to several factors, including: A decrease in the amount of water compared to vegetative cells An increase in the amount of dipicolinic acid and calcium ions Enzymes which are more resistant to heat A spore coat which is impermeable to many substances Endospores may be formed in a central, terminal, or sub-terminal position in the cell and their shape varies from ellipsoidal to spherical. The location of the endospore in the cell is usually characteristic of the species. For example, the location and shape of the Bacillus subtilis endospore is different from the location and shape of the 57 Clostridium endospore. Therefore, the presence or absence of endospores and the description of the endospore is useful to a microbiologist as an aid in identification. The resistant properties of endospores make them difficult to stain, hence heat is used in conjunction with staining to enable the stain to penetrate into the spore coat. Preparation of Films for Staining – Procedure Obtain a clean slide and draw a circle on it approximately 1.5 cm in diameter. Turn the slide over. Flick the tube of culture to mix up the cells, and use a loop to obtain aseptically a drop of culture. Place this loopful of culture within the circle. Alternatively, if using a plate culture, first use your loop to add a drop of water to the circle on the slide. Remove a small quantity of culture and mix with the water to make a smooth suspension. Allow the suspension to air dry. When dry, the film should be only faintly visible; a thick opaque film is useless. The only fixation required is to pass the slide several times (maximum 10) through the bunsen burner flame until the slide is warm but not too hot. If the slide is fixed until too hot to the touch, the bacteria will be misshapen when observed under the microscope. Gram Staining – Procedure Prepare smear, dry and heat fix. Flood the smear with crystal violet solution for 1 min. Gently wash with tap water for 2-3 seconds and remove the water by tapping the slide gently on paper towel. Add Gram’s iodine solution to the slide for 1 min. Wash gently with tap water and remove as above. Decolourise with 95% ethanol by dripping ethanol on surface of slide until no more colour is removed. Rinse gently with water. If too much alcohol is added, the Gram-positive organisms may become Gram-negative. Remove the water after the last wash. Counterstain the slide with safranin for 30 seconds - 1 minute. Wash the slides with tap water, air dry on paper towels, and examine under oil immersion. Gram positive organisms stain purple; Gram negative organisms, red (pink). Negative Staining – Procedure Add a small drop of India ink to a wet mount of bacterial cells on a microscope slide. Mix with a toothpick. 58 Add a coverslip, place the slide between two sheets of blotting paper, and apply pressure with a cork until most of the India ink is removed (slide should appear a light gray in colour) Observe using phase contrast microscopy. PHB Staining - Procedure Prepare smears of the organism, air dry and heat fix. Flood entire slide with Sudan Black B and add more stain as the dye solvent evaporates. Stain for at least 10 minutes. Pour off excess stain (do not wash) and air dry. Clear slide by dipping in a jar of solvent in the fume hood for 5 sec. Air dry in the fume hood. Counterstain for 1 min. with safranin. Wash with water, drain, blot and air dry. Examine with oil immersion objective. Cell is pink, lipids are dark grey or black. Endospore Staining - Procedure Prepare smear and heat fix. Cover the dried fixed film with a small piece of paper towel. Saturate this with 5% malachite green. Place the slide on a rack over a boiling water bath. Steam slide for 5-10 minutes in this manner. Add additional stain as needed - do not allow the slide to dry out during this procedure. Caution: the water bath is at 100oC – the steam will burn your skin. Take appropriate precautions. Allow the slide to cool, then rinse with water. Tap over a paper towel to remove excess water Counterstain with safranin for 30 seconds. Rinse slide with water. Allow to air dry, and view. Endospores will stain green and the rest of the cell pink. References: Atlas, R. M. 1997. Principles of Microbiology. Wm. C. Brown Publishers, Toronto. Madigan, M. T., and Martinko, 2006. Brock Biology of Microorganisms Eleventh Edition. Prentice-Hall of Canada, Inc., Toronto. Ross, H. 1992-1993. Microbiology 241 Laboratory Manual. The University of Calgary Press, Calgary. 59 APPENDIX 4 ASEPTIC TECHNIQUE A. Aseptic Technique Much microbiological work, and to some extent biochemical work, depends on the maintenance of pure cultures of microorganisms. Therefore, there are various essential precautions that MUST be observed to exclude unwanted organisms. Accidental contamination may ruin your results completely. Aseptic technique is largely a matter of common sense, but it is essential to realise that bacterial and fungal spores are present everywhere, and a high standard of technique must be attained. Correct methods of handling cultures and apparatus will be demonstrated. methods must be followed. These Consider carefully and remember the following points: 1. Clean air contains many bacterial and fungal spores carried on dust particles or in water droplets. Any surface exposed to air quickly becomes contaminated, and if material is to be kept sterile it should be exposed only as much as is absolutely necessary for manipulation. Instruments which can be sterilised by heating in a bunsen flame (e.g. inoculating loops) can be left exposed, but they must be flamed thoroughly before use, and again before being replaced in the holder. Items of equipment that cannot be treated in this way (e.g. pipettes) are sterilised in wrappings or containers from which they must not be removed until actually needed. They must not be allowed to touch unsterile surfaces during use. Plugs and caps of tubes and bottles must not be laid on the bench nor must sterile containers be left open to collect falling dust. 2. Clothes, hair, skin and breath all carry a heavy microbial load and where strict asepsis is essential, sterilized gowns, caps, gloves etc. are worn. Even in normal microbiological work care must be taken to prevent contamination from the above mentioned sources. A clean laboratory overall is advised for all lab work. Microbial contamination in the lab is most often due to currents of unsterile air. The chief merit of inoculation chambers and screens therefore lies in the protection they give from drafts. This protection can be supplemented by keeping all windows and doors shut and by cutting down personal movement within the laboratory. These precautions can be offset by careless use of burners that create convection currents. 60 3. Before any operation is started, all necessary materials should be assembled in convenient order with provision for protecting sterile objects until needed, and for disposing of used apparatus (so as not to contaminate other material). B. Aseptic Culture Manipulation Purposes: 1) To prevent the contamination of the environment and people working in the laboratory from the cultures used in the exercises 2) To prevent accidental contamination of cultures of microorganisms and of solutions and equipment used in the laboratory Correct methods of handling cultures and apparatus will be demonstrated. These methods must be followed. Consider carefully and remember the following points: Prior to starting any work in the laboratory, wash hands with soap, and wash down bench area using 10% bleach. This procedure should be repeated after the lab is complete. Avoid working on your lab book or lab notes. Clean laboratory coats must be worn. If you have long hair, tie it back before working in the laboratory environment. Eating or drinking is not permitted in the laboratory. Do not place pencils, fingers or anything else in your mouth. Clean air contains many bacteria and fungal spores carried on dust particles or in water droplets. Any surface exposed to air quickly becomes contaminated. If material is to be kept sterile, it should be exposed only as much as is absolutely necessary for manipulation. Plugs and caps of tubes, tops of Petri dishes and bottles of solutions, (even water!!) must not be laid on the bench nor must sterile containers and cultures be left open and exposed to the air. Inoculation of Culture Tubes Again, the important thing to remember is that exposure of sterile liquids or bacterial cultures to air must be minimised. -Ensure that you have the tubes, plate of inoculum, inoculating loop and a sterile tube of medium available within easy reach. 61 -Flame the inoculating loop until red hot. When removing inoculum from a tube, remove the cap from the tube by grasping the cap between the last finger and the hand which is also holding the inoculating needle (Figure 1). Do not place the cap on the bench!! Figure 1: Technique for manipulating test tubes aseptically. -Flame the mouth of the tube by passing it rapidly through the Bunsen burner 2-3 times. This sterilises the air in and immediately around the mouth of the tube. -Cool the loop on the inside of the tube, remove the inoculum. -Reflame the mouth of the tube and replace the cap -Flame the inoculating loop before replacing -Note, when removing inoculum from a plate, cool the loop in the agar before picking up the bacteria Streaking for Single Colonies -A loop of liquid culture or a small amount of bacterial growth from a plate culture is transferred aseptically to a sterile plate in the area shown by Figure 2A. -Once the first set of streaks has been made, the inoculating loop is reflamed until red hot. DO NOT REINTRODUCE THE LOOP INTO THE ORIGINAL CULTURE!!! -Cool the loop, and make a second set of streaks as shown in Figure 2B, only crossing over the initial set of streaks once. -Flame the loop again, cool, and repeat for three more sets (Figure 2C). Note, try not to gouge the agar while streaking the plate. 62 Figure 2: How to prepare a streak plate. Preparation of Spread Plates: Generally, volumes of culture greater than 100 L are NOT plated as it takes too long for the liquid to dry. Use aseptic technique to obtain 100 L of culture and place in the middle of a plate of medium. Use a sterile glass spreader (this may involve dipping a spreader into a beaker of alcohol and waving it through a Bunsen Burner flame. If this is the case, DO NOT hold the spreader in the flame and avoid tipping the spreader so that flaming alcohol runs over your hand. Once the flame has burnt out, the spreader is ready to use). Use the same hand that holds the spreader to lift the lid of the plate and keep it just above the plate the entire time. 63 Gently touch the spreader to the side of the medium (not directly in the culture in case the spreader is still a bit warm). Smooth the culture evenly over the surface of the plate ensuring that you cover the entire plate. Invert the plates and place in the incubator when dry C. Sterilization Media must be sterilised after distribution into tubes, flasks or bottles. Sterilised media may later be transferred aseptically to previously sterilised containers, but this should only be done when really necessary, e.g. in preparing "plate" cultures, since some risk of contamination is unavoidable. Methods of Sterilization 1. Most media (including agar) can be sterilised by treatment with steam under pressure in an autoclave, the usual treatment being 15-20 minutes at a pressure of two atmospheres. This raises the steam temperature to 121°C. When using an autoclave, the water should be allowed to boil, and the steam to fill the autoclave before shutting the valve. This allows the material to heat up and ensures that the correct steam pressure is attained. Never overfill an autoclave since this will upset the pressure/volume relationship and the correct temperature will not be attained. Materials that might be adversely affected by this treatment may sometimes be treated for a short time or at a lower temperature, but this will not be effective if the material is heavily contaminated to begin with. Screw caps on bottles must be left slightly open during sterilisation and screwed down on removal from the steriliser. 2. Media that are difficult or impossible to autoclave satisfactorily, e.g. gelatin media and some sugar media, may be sterilised by intermittent steaming. Objects to be sterilised are heated over boiling water in a steamer (steam temperature 85°-95°C) for 15-20 minutes on each of three or more successive days. Time must be allowed for the medium to reach the same temperature as the steam. Between treatments the material must be kept at a temperature allowing spores to germinate (30°-37°C) and so lose their heat resistance. 3. It is often necessary to sterilise some ingredients of a medium separately and to add them to the rest of the medium before use. Heat-labile ingredients, e.g. urea, serum, etc. must be sterilised by filtration through a bacteria-proof filter, i.e. Seitz filters or membrane filters. 4. Dry glassware, e.g. glass petri dishes, empty flasks, pipettes may be sterilised in the autoclave and then dried or may be sterilised in a hot air oven. Any oil material must also be sterilised in a hot air oven. The minimum effective treatment is 1 hour at 150°C. This should be increased to 160°C or the time of heating prolonged to 2 or 3 hours wherever possible. 64 APPENDIX 5 THE CULTIVATION OF BACTERIA In order to grow, microorganisms require a) water, b) macronutrients eg. – C, N, K, P, S, Mg, Ca, Na, and Fe c) micronutrients (trace elements) eg. - Fe, W, Zn and d) growth factors – vitamins, amino acids, purines and pyrimidines. In general, wild-type organisms are termed prototrophs. An auxotroph is a nutritional mutant, unable to synthesise an essential component for growth from precursors. Note that this essential component is normally synthesised by the wild-type or prototrophic strains of the same species. Scientists study and manipulate nutritional requirements of bacteria or yeast using minimal media. Minimal or defined media are those in which the exact chemical composition of all ingredients is known. A medium where the exact chemical composition is not known is termed complex. Complex media are preferred as they are generally easier to prepare than minimal media, they result in high levels of growth, and are useful when exact nutritional requirements of an organism are not known. Nutritional Classification: The nutritional classification of organisms is based on three parameters: the energy source, the principal carbon source and the source of reducing power. With respect to energy source, phototrophs are photosynthetic organisms that use light as their energy source and chemotrophs are organisms that depend on a chemical energy source. Organisms able to use CO2 as a principal carbon source are autotrophs. Heterotrophs depend on an organic carbon source. To designate the source of reducing power, the term lithotroph or organotroph is applied. Lithotrophs use inorganic compounds as their source of reducing power, and organotrophs use organic compounds as their source of reducing power. To summarise: photoautotroph (photolithotroph) photoheterotroph (photoorganotroph) chemoautotroph (chemolithotroph)* chemoheterotroph (chemoorganotroph) energy source light carbon source CO2 light organic chemical (oxidation of CO2 reduced inorganic compounds e.g. NH3, NO2- and H2) chemical organic 65 source of reducing power inorganic oxidizable substrate organic inorganic organic *All chemoautotrophs are chemolithotrophs, but not all lithotrophs are autotrophic. For example, the methylotrophic bacteria can use organic carbon as their carbon source. Common Media Constituents: Energy or Carbon sources: Sugars, alcohols, carbohydrates and amino acids Found in infusions – for instance – beef infusion Found in extracts – for instance – yeast extracts Also found in peptones (see below) Nitrogen sources: Inorganic sources such as ammonia or nitrate Nitrogen fixing organisms use atmospheric N2 Extracts, infusions Peptone – hydrolysis of proteins produces mixtures of short-chains of amino acids (peptides). Sources of peptones may include meat, fish, blood, or soybeans Tryptone – pancreatic digestion of casein Other Macronutrient Source Examples: MgSO4 CaCl2 Potassium salts Micronutrient Sources: May not be necessary to add as these are required in such small concentrations. Growth Factors: Some organisms are able to synthesise all growth factors from precursors. Other organisms require these compounds already synthesised For example – thiamine, biotin Buffering Components Buffers, which prevent large changes in pH, are often required to facilitate growth. This is particularly true of media composed of simple compounds or in which acid-producing bacteria are cultivated. Mixtures of sodium and potassium phosphates are often employed. In complex media, buffering is provided by the peptides and amino acids. Gelling Agents For a solid medium, agar, a water soluble polysaccharide, is added to the medium. First discovered in 1658 in Japan, agar was first used for microbiological purposes by R. Koch in 66 1882. It is extracted from members of Class Rhodophyceae (a group of red-purple marine algae). Agar is particularly suited to microbial propagation because: It lacks metabolically useful chemicals such as peptides and fermentable carbohydrates (it cannot be broken down by bacterial enzymes) It melts at a high enough temperature (85 oC) to support growth of different temperature requiring microbes It lacks bacterial inhibitors Below are two examples of media used for cultivation of microbes. TY is an example of a complex medium whereas VMM is an example of a minimal or defined medium: TY Agar (used for the cultivation of organisms such as Rhizobium leguminosarum, Pseudomonas fluorescens) As with most complex media, ingredients for TY are weighed out, 1 L of water is added, and the mixture autoclaved. After cooling slightly to approximately 60 oC, TY medium is poured into Petri dishes. Ingredient Tryptone Amount (/L) 5.0 g Yeast Extract 3.0 g CaCl2 MgSO4 Agar 0.5 g 0.1 g 20 g Source of? Macronutrients (primarily nitrogen, also carbon and growth factors in the form of amino acids) Macronutrients (primarily carbon, also nitrogen and growth factors) Macronutrients Macronutrients Gelling agent For the next example – VMM – three different mixtures (Solutions A, B and C) of ingredients are made up separately, autoclaved separately, and then combined. Finally, a carbon source is added just prior to pouring. 67 VMM (Vincent’s Minimal Medium - Vincent, 1970) (used for the study of nutritional requirements of Rhizobium leguminosarum) Solution A: Source of? Compound Amount (/L) Buffering agent/ K2HPO4 1.0 g Macronutrients Buffering KH2PO4 1.0 g agent/Macronutrients Macronutrients (nitrogen KNO3 0.6 g in particular) Gelling agent For Solid Medium: 12.5 g Agar Solution B (10x): Compound FeCl3 Amount (/L) 0.1 g Source of? Macro/Micronutrient s MgSO4 2.5 g Macronutrients CaCl2 1.0 g Macronutrients Autoclave and add to a final concentration of 1x Solution C (100x) Compound Amount for: 1 L Source of? Biotin 0.01 g Growth factors Thiamine 0.01 g Growth factors Calcium 0.01 g Growth factors Pantothenate Autoclave and add to a final concentration of 1x. Carbon sources: Depending on the organism studied, a variety of carbon sources may be added. For instance, when studying genes required for catabolism of a certain carbon source, a scientist will often first create a mutant or auxotroph unable to catabolise that carbon source. To confirm presence of the mutation, it is necessary to plate the putative auxotroph on medium containing the carbon source of interest, and plating on a medium containing a carbon source that the organism is able to utilise. In Rhizobium leguminosarum, some examples of carbon sources that are useful for these types of experiments are mannitol, sorbitol (both are sugar alcohols), or rhamnose. Each carbon source is prepared as a stock solution, filter sterilised, and added to a final concentration of 0.4% (w/v). 68 Oxygen Requirements of Microorganisms Many species of bacteria are facultative aerobes, i.e. they can grow under aerobic or anaerobic conditions, the latter ability being dependent upon the presence of some substance that can be utilised as an electron acceptor by the species concerned. Some bacteria are obligate aerobes, unable to use anything but oxygen as a final electron acceptor. Others are obligate anaerobes that cannot use oxygen as an electron acceptor. A few bacteria are somewhat intermediate, growing best in low oxygen tensions. These are called microaerophilic bacteria. During growth in liquid culture, microorganisms tend to utilise all available oxygen and so reduce the medium. Thus, the oxidation-reduction potential (Eo) of the medium may become low enough to allow anaerobic growth to occur. One example of this is found in the fermentation of sugar to produce alcohol by yeast (Exercise 8 part C). Unless the mixture is stirred frequently, the little oxygen available in the grape juice solution is utilised rapidly by the growing culture. Organisms then switch to anaerobic growth. In order to sample material containing anaerobes, specimens must be obtained and immediately placed into an environment containing an oxygen-free gas and an indicator that changes colour when oxidised to indicate when oxygen has contaminated the sample. Organisms may then be cultured in sealed jars containing gas mixtures of N 2 and CO2 or even by cultivation in an anaerobic chamber. Temperature Requirements of Microorganisms Cultures should be incubated at the temperature most favourable to growth or the specific activity being studied. Human pathogens and commensal species grow best at body temperature, i.e. 37°C. Soil organisms and plant pathogens are normally incubated at 20-30°C. The optimum temperature is that temperature at which the growth rate is maximal for a particular organism. Note that for every organism, there is also a minimum temperature below which no growth occurs, and a maximum temperature, above which no growth occurs. The terms used to describe microorganisms according to their temperature requirements are as follows: thermophiles require temperatures of 45°C-65°C extreme thermophiles (which are usually archaebacteria) will grow at temperatures above 65°C. mesophiles grow best at temperatures of 20°C-45°C. psychrophiles require low temperatures - below 15°C. 69 References: Difco Manual. 1998. Difco Laboratories, Division of Becton Dickinson and Company, Maryland. Madigan, M. T., Martinko, J. M., and Parker, J. 2003. Brock Biology of Microorganisms 10th Edition. Prentice-Hall Canada Inc., Toronto. Ross, H. 1992/3. Microbiology 241 Lab Manual. University of Calgary Press, Calgary. 70 APPENDIX 6 BACTERIAL OBSERVATION Bacterial genera may be differentiated in two ways: 1) by the cellular morphology which is observed microscopically 2) by the colony morphology which is observed on a plate culture Cellular Morphology includes: 1) Shape: rods, cocci, spirilli 2) Size (in m): diameter (cocci); lengthxwidth (rods) 3) Typical arrangement of the cells: chains, clusters, pairs, random 4) Gram reaction A diagram drawn to scale accompanies the cellular morphology. Colony Morphology is that of a single isolated colony on the plate, not the morphology of the entire bacterial growth on the plate. Colony morphology is influenced by medium composition; type of medium organism is grown on (defined, complex, specific type) should be noted in conjunction with the description of colony morphology. The following characteristics are those most commonly used to describe colony morphology: 1) Shape or form Circular Irregular RhizoidFilamentous Punctiform (1mm or less in diameter) 2) Surface: smooth/rough; mucoid/moist/dry/powdery 3) Elevation: Flat 4) 5) 6) Raised Convex Umbonate Umbilicate Size: measure a single colony with a ruler Pigment: cream, white or beige coloured organisms are usually considered to be non-pigmented. Pigments may be purple, red, pink, yellow, brown, blue, grey, etc. Water soluble pigments diffuse into the medium. Opacity: Transparent (can see through) or opaque. 71 APPENDIX 7 LABORATORY REPORTS Lab reports shall be in the style of scientific papers published in refereed journals. This scientific style is relatively similar across journals although specific formats vary, including the form of literature citations. The journals Microbiology or Canadian Journal of Microbiology will be used as models for the specific format of Biology 3200 reports. Please do not use formats from journals such as Nature or Proceedings of the National Academy of Sciences as this will result in loss of marks. For detailed information on preparation of scientific reports, please refer to the Biology 3200 web site. The text should be in prose form and standard rules of grammar apply. Check spelling, including technical terms and names of bacterial species which are italicised or underlined; for instance, Escherichia coli or Escherichia coli. The reports shall be double-spaced, single-sided and typed. Staple the report together and do not submit it in a cover. The reports shall contain the normal components of a scientific paper including: Title - the title should identify the experimental topic as completely as possible. Abstract - the abstract is an abbreviated version of the complete report. Typically containing no more than 250 words, the abstract picks out the highlights of the introduction, methods, results and discussion. The abstract should be complete enough that it can be removed from the report and will still provide a meaningful description of the study. Introduction - The introduction serves to (i) provide background information and a description of what is known prior to the study, and (ii) offer a justification for the study. This justification describes why the experiment was performed - how does it fit into science and are there any applied aspects of the knowledge (i.e. is it relevant to medicine, agriculture or other disciplines). Relevant literature is used and cited. Methods - The methods or 'Materials and Methods' describes the materials involved in the study, including biological materials (bacteria, etc.), and outlines the procedures used in the study. Reference must be made to this laboratory manual (Pacarynuk and Danyk, 2004). Other references, the text by Madigan et. al., (2003), or other published materials may be cited. Global referencing (“All of the following methods are taken from…”) should be avoided. The methods section should be adequate for the reader to completely understand what was done and also to be able to repeat fully the study. Results - The results describe the observations or experimental outcomes, providing figures, tables or other data as suitable. This section answers the question “What Happened?” The author should decide what is the most suitable format for experimental 72 information and draft the report accordingly. Figures may include drawings that should be in pencil. Graphs or other figures may also be included as appropriate. Experimental results should be presented only once. If information is presented in a figure then it should not be repeated in a table. Each figure and table must have a caption which is complete enough that the figure and caption can be removed from the report and still be understandable. Figures and tables must be referred to in the text and described so that if the reader did not have the figure or table, trends or highlights of the results would still be evident. Never include a figure or table without referring to it and describing it; to do so will result in loss of marks. Avoid evaluating or interpreting your results in this section. Discussion - The discussion should refer to concepts or questions posed in the introduction and relate these concepts from the literature to the results. Do not restate the results in this section. Your discussion will be graded based on your evaluation of the results with respect to the literature. Any time you use information from another source, it must be immediately cited within the text. Failure to do this constitutes plagiarism and may result in a mark of zero being assigned for the entire document. For examples of how to cite properly, refer to peer-reviewed journal articles in Microbiology or Canadian Journal of Microbiology. Never include quotations, such as phrases from the course text or this lab manual. Direct quotes are inappropriate in scientific writing. Always introduce relevant concepts using your own wording and then cite using the format found in Microbiology or in Canadian Journal of Microbiology. Literature Cited – This section only includes references cited within the body of the text. Again, use the format found in Microbiology or the Canadian Journal of Microbiology. References will include journal papers, books and most likely, Holt (1989) or Holt (1994) (Bergey’s Manual of Systematic Bacteriology). It is important to note that Bergey did not write Bergey’s Manual of Systematic Bacteriology; the proper formats for referencing are as follows: Holt, J. G. (editor-in-chief). Bergey’s Manual of Systematic Bacteriology, Vol. I, 1984; vol. II, 1986; vols, III and IV, 1989. Williams and Wilkins, Baltimore. Holt, J. G. (editor-in-chief) (1994). Bergey’s Manual of Determinative Bacteriology, 9th edition. Williams and Wilkins, Baltimore. 73 APPENDIX 8 USE OF THE SPECTROPHOTOMETER Many procedures for the quantitative analysis of compounds in biological fluids are based on the fact that such compounds will selectively absorb specific wavelengths of light. For example, a solution that appears red to us (such as blood) absorbs the blue or green colours of light, while the red is reflected to our eyes. The eye, however, is a poor quantitative instrument, and what appears bright red-orange to one person may appear dull red-purple to another. A spectrophotometer is one instrument that will objectively quantify the amount and kinds of light that are absorbed by molecules in solution. A source of white light is focused on a prism to separate it into its individual bands of radiant energy (Figure 1). One particular wavelength is selected to pass through a narrow slit and then through the sample being measured. The sample, usually dissolved in a solvent, is contained in an optically selected tube or cuvette, which is standardized for wall thickness and has a light path exactly one centimeter across (these tubes are therefore expensive!). Figure 1. A photoelectric spectrophotometer. After passing through the sample, the selected wavelength of light strikes a photoelectric tube. If the substance in the cuvette has absorbed any of the light, the light transmitted out the far side will then be reduced in total energy content. When it hits the photoelectric tube, it generates an electric current proportional to the intensity of the light energy striking it. By connecting the photoelectric tube to a device that measures electric current (a galvanometer), a means of directly measuring the intensity of the light is achieved. The galvanometer has two scales: one indicates the % transmittance, and the other, a logarithmic scale with unequal divisions graduated from 0.0 to 2.0, indicates the absorbance. 74 Zeroing the Spectrophotometer Because most biological molecules are dissolved in a solvent before measurement, a source of error can be the absorption of light by the solvent. To assure that the spectrophotometric measurement will reflect only the light absorption of the molecules being studied, a mechanism of "subtracting" the absorbance of the solvent is necessary: 1) Align the needle to 0 on the transmittance scale using the knob on the left hand side of the machine (as you face the machine). Note, this step should be performed prior to placing any tubes into the machine. 2) Insert the reagent "blank" (the solvent) into the instrument, and align the needle to 0 on the absorbance scale using the knob on the right hand side of the machine (as you face the machine). 1) The sample, containing solute plus solvent, is then inserted. Any reading on the scale that is less than 100% transmittance (or greater than 0.0 absorbance) is considered to be due to absorbance by the solute only. Units of measurement: The transmittance scale is a % number; a ratio of the light exiting the sample tube to the light entering the tube. However, this number is not a linear reflection of the concentration of the solute molecules (Figure 2). The absorbance scale, on the other hand, does reflect a linear relationship. Although you do not necessarily know the exact concentration of the solute molecules in your sample, you do know that if the absorbance value doubles, the concentration of solute in your sample has doubled. Absorbance has no units, but the wavelength of the light is usually indicated by a subscript. Figure 2. The relationship between % transmittance and solute concentration (on the left), and absorbance and solute concentration (on the right). 75 APPENDIX 9 Media, Reagents and pH Indicators MEDIA: Tryptic Soy Broth: A general purpose medium used to cultivate a variety of microorganisms. Composition (g/L): Bacto tryptone Bacto soytone Dextrose NaCl Dipotassium phosphate 17.0 g 3.0 g 2.5 g 5.0 g 2.5 g Dissolve in distilled water to a final volume of 1 L, dispense into test tubes, and autoclave for 15 min at 121oC. Tryptic Soy Agar: Used for cultivation of a variety of microorganisms. Composition (g/L): Bacto tryptone Bacto soytone NaCl Agar 15.0 g 5.0 g 5.0 g 15.0 g Dissolve in distilled water to a final volume of 1 L, autoclave for 15 min at 121 oC, and pour into sterile Petri dishes. LB Medium (Luria-Bertani Medium): Used for cultivation of Enterobactereaceae family members, Sinorhizobium and Agrobacterium Composition (g/L): Tryptone 10.0 g Yeast extract 5.0g NaCl 10.0 g Dissolve in distilled deionised H2O to a final volume of 1 L, autoclave for 20 minutes at 15 psi (1.05 kg/cm2) on liquid cycle, and pour into sterile Petri dishes. 76 Terrific Broth (TB) Used for the cultivation of E. coli Composition (g/L) Tryptone Yeast Extract Glycerol 12.0 g 24.0 g 4.0 mL Dissolve in distilled deionised H2O to a final volume of 900 mL, autoclave for 20 minutes at 15 psi (1.05 kg/cm2) on liquid cycle. Allow the solution to cool to 60 oC or less, and then add 100 mL of a sterile solution of 0.17M KH PO , 0.72M 2 4 K2HPO4 (this is the solution resulting from dissolving 2.31 g of KH2PO4 and 12.54g of K2HPO4 in 90 mL of deionised H2O. After the salts have dissolved, adjust the volume of the solution to 100 mL with deionised H2O and sterilise by autoclaving for 20 minutes at 15 psi on liquid cycle). Nutrient Agar: Used for the cultivation of a wide variety of microorganisms. Composition (g/L) Peptone NaCl Yeast extract Beef extract Agar 5.0 g 5.0 g 2.0 g 1.0 g 15.0 g Dissolve in distilled water to a final volume of 1 L, autoclave for 15 min at 121 oC, and pour into sterile Petri dishes. TY Agar Used for the cultivation of Pseudomonas and Rhizobium. Composition (g/L): Tryptone Yeast Extract CaCl2 MgSO4 Agar 5.0 g 3.0 g 0.5 g 0.1 g 13.0 g 77 Add distilled water to a final volume of 1 L, autoclave for 15 min. at 121 oC, and pour into sterile Petri dishes. Luria Methylene Blue Agar Used for the observation of coliphage plaques. Composition (g/L): Tryptone Yeast Extract NaCl Glucose Methylene Blue Agar 10.0 g 5.0 g 5.0 g 1.0 g 0.02 g 15.0 g Dissolve in distilled deionised H2O to a final volume of 1 L, autoclave for 20 minutes at 15 psi (1.05 kg/cm2) on liquid cycle, and pour into sterile Petri dishes. Luria Agar Overlay Used for the propagation of coliphage. Composition (g/L): Tryptone NaCl Glucose CaCl2 Agar 10.0 g 5.0 g 1.0 g 0.11 g 6.0 g Add 3 mL of NaOH per L and check for a pH of 7.2. Add agar, dissolve, then autoclave for 20 minutes at 15 psi (1.05 kg/cm2) on liquid cycle. 78 Eosin Methylene Blue Agar: Used for selection of Gram negative bacteria, and differentiation of lactose fermenting organisms. Composition (g/L): Peptones Di-potassium hydrogen phosphate Lactose Sucrose Eosin Y, yellowish Methylene blue Agar 10.0 g 2.0 g 5.0 g 5.0 g 0.4 g 0.07 g 15 g Dissolve in distilled water to a final volume of 1 L, autoclave 15 min at 121oC, and pour plates. MacConkey Agar: Used for selection of Gram negative bacteria, and differentiation of lactose fermenting organisms. Composition (g/L): Peptone NaCl Lactose Bile salts Neutral red Agar 20.0 g 5.0 g 10.0 g 5.0 g 0.075 g 12.0 g Dissolve in distilled water to a final volume of 1 L, autoclave 15 min at 121 oC, and pour plates. 79 References: Atlas, R.M., and Parks, L.C. 1993. Handbook of Microbiological Media. CRC Press, Inc. Boca Raton, Florida. Difco Manual: Dehydrated Culture Media and Reagents for Microbiology. 10th Ed. (1984). Difco Laboratories, Detroit, Michigan. Merck Microbiology Manual 1994. Merck, Darmstadt, Germany. Ross, H. 1992/3. Microbiology 241 Lab Manual. University of Calgary Press, Calgary. Sambrook, J. and Russell, D. W. 2001. Molecular Cloning – A Laboratory Manual. 3rd edition. Cold Spring Harbor Laboratory Press, New York. 80 REAGENTS: Ethanol, 70%: 95% Ethanol Distilled Water 36.8 mL 13.2 mL Barritt’s Reagents: Solution A: Dissolve 6 g alpha naphthol in 100 mL 95% ethanol Solution B: Dissolve 16 g potassium hydroxide in 100 mL distilled water. Crystal Violet Stain: Solution A: Dissolve 2.0 g of crystal violet in 20 mL of 95% ethanol. Solution B: Dissolve 0.8 g of ammonium oxalate in 80 mL of distilled water. Mix solutions A and B. Gram’s Iodine: Dissolve 2 g of potassium iodide in 300 mL of distilled water; then add 1 g of iodine crystals. Kovac’s Reagent: Mix the following: n-Amyl alcohol Hydrochloric acid p-dimethylamine-benzaldehyde 75 mL 25 mL 5.0 g Malachite Green Stain: Dissolve 5 g of malachite green oxalate in 100 mL of distilled water. Nigrosin Solution: Add 10 g of nigrosin (water soluble) to 100 mL of distilled water. Boil for 30 min, and add 0.5 mL of formaldehyde (40%). Filter twice through double filter paper. Store under aseptic conditions. Oxidase Test Reagent: Dissolve 1 g of dimethyl-p-phenylenediamine hydrochloride in 100 mL of distilled water. Make fresh. Phloxine B: Dissolve 1 g of phloxine in 100 mL of distilled water. Safranin: Dissolve 0.25g safranin in 10 mL of 95% ethanol. Add to 100 mL of distilled water. 81 Sudan Black Stain: Dissolve 0.3 g of Sudan Black in 100 mL of 70% ethanol. Shake before each use. References: Clark, G. (1984) Staining Procedures. 4th Ed. Williams and Wilkins, Baltimore, Maryland. Benson, H.J. (1985). Microbiological Applications: A Laboratory Manual in General Microbiology, 4th Ed. Wm. C. Brown Publishers, Dubuque, Iowa. 82 pH INDICATORS: Table 1: Indicators of Hydrogen Ion Concentration. pH Indicator pH Range Full Acid Colour Full Alkaline Colour Cresol Red 0.2 - 0.8 Red Yellow Meta Cresol Purple (acid range) Thymol Blue 1.2 - 2.8 Red Yellow 1.2 - 2.8 Red Yellow Brom Phenol Blue 3.0 - 4.6 Yellow Blue Brom Cresol Green 3.8 - 5.4 Yellow Blue Chlor Cresol Green 4.0 - 5.6 Yellow Blue Methyl Red 4.4 - 6.4 Red Yellow Chlor Phenol Red 4.8 - 6.4 Yellow Red Brom Cresol Purple 5.2 - 6.8 Yellow Purple Bromothymol Blue 6.0 - 7.6 Yellow Blue Neutral Red 6.8 - 8.0 Red Amber Phenol Red 6.8 - 8.4 Yellow Red Cresol Red 7.2 - 8.8 Yellow Red Meta Cresol Purple (alkaline range) 7.4 - 9.0 Yellow Purple Thymol (alkaline range) Blue 8.0 - 9.6 Yellow Blue Cresolphthalein 8.2 - 9.8 Colourless Red Phenolphthalein 8.3 - 10.0 Colourless Red Adapted from: Benson, H.J. (1985). Microbiological Applications: A Laboratory Manual in General Microbiology, 4th Ed. Wm. C. Brown Publishers, Dubuque, Iowa 83