Chromosomes and chromatin
advertisement

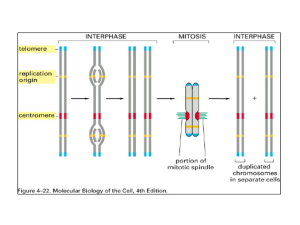
Chromosomes and chromatin Chromosomes organize and package genes inside cells • Bind packaging proteins to DNA to make it more compact. – Histones +DNA = chromatin in eukaryotes – Virion proteins in viruses – HU (?) or other proteins in bacteria • Loop chromatin and attach it to a matrix in nuclei Bands and specialized regions of human chromosomes TEL 15 .5 15 .4 .3 15 .2 15 15 .1 short arm (p) Centromere = CEN p1 5 p1 4 p1 3 p1 2 p1 1 HRAS HBB PTH MYOD long arm (q) Geimsa dark band ATA=ataxia telangiectasia Geimsa light band Telomere = TEL (CCCTAA) n or (AGGGTT) n Human chromosome 11: 125 Mb, 180 cM Human chromosomes, ideograms Mitotic chromosomes are spread and stained with Geimsa. Those that stain are shown in black. G-bands (more A+T rich). Human chromosomes, spectral karyotype Reagents specific to each chromosome. Chromosome painting. Identifying translocations http://www.ncbi.nlm.nih.gov/disease/ Distinctive and common features of chromosomes • Distinctive proteins and DNA sequences have been used to develop chromosome painting reagents. • Genomic DNA in vertebrates has long (megabase) stretches of G+C rich DNA, and other long stretches of A+T rich DNA – Called isochores • Virtually all this DNA is organized into chromatin, which has a common fundamental structure. Chromatin Structure Principal proteins in chromatin are histones H3 and H4 : Arg rich, mostly conserved sequence H2A and H2B : Slightly Lys rich, fairly conserved H1 : very Lys rich, most variable in sequence between species Histone structure and function Histone structure and function "Minimal" structure for a core histone, e.g. H4. Others have one additional alpha helix. N K5 K8 K12 K16 Highly charged N-terminal tail. L1 L2 C Globular, hydrophobic domain for histone-histone interactions and for histone-DNA interactions. Histone interactions via the histone fold The alpha-helical regions of the core histones mediate dimerization. N L2 L1 L2 C C C L1 L1 N The histone fold flanked by N and C terminal tails. L2 N Dimer of histones joined by interactions at the histone fold. Nucleosomes are the subunits of the chromatin fiber • Experimental evidence: – Beads on a string in EM – Micrococcal nuclease digestion General model for the nucleosomal core A string of nucleosomes Detailed structure of the nucleosomal core Higher order chromatin structure Histone H1 associates with the linker DNA, and may play a role in forming higher order structures. Alterations to chromatin structure are key steps in regulation Phosphorylation of histones Serines in histones can be modified by phosphorylation O -O protein kinase ATP OH O CH 2 ... NH CH C NH CH C ... 2 Gly Ser O phosphate H 2O protein phosphatase OH O ADP O P O CH 2 ... NH CH C NH CH C ... 2 Gly Ser Negative charge on phosphoserine mammalian chromatin asssembly factor 1 (CAF1) Ac Ac Acetylation and Deacetylation of K5 K8 K12 K16 lysines in proteins + NH 3 CH 2 CH 2 AcCoA O CH 2 O CH 2 ... NH CH C NH CH C ... 2 Gly Lys Ac Positive charge on amino group CH 3 O C NH CH 2 CH 2 CoA O CH 2 O CH 2 ... NH CH C NH CH C ... 2 No charge on amide group Acetylation and Deacetylation of Histone acetylation and deacetylation are key processes in chromatin assembly and regulation histones E.g., Histone H4 Ac Ac K5 K8 K12 K16 Histone deacetylases, e.g. yeast Rpd3p and mammalian HD1 Nuclear histone acetyl transferase (HAT A), e.g. yeast Gcn5p and its mammalian homolog PCAF N L1 L2 C K5 K8 K12 K16 Histone deacetylases Cytoplasmic HAT B, e.g. yeast Hat1p + Hat2p and mammalian chromatin asssembly factor 1 (CAF1) Ac Ac K5 K8 K12 K16 + NH 3 O CH 3 C Effects of histone modifications • Highly acetylated histones are associated with actively transcribed chromatin – Acetylation of histone N-terminal tails may affect the ability of nucleosomes to associate in higher-order structures – The acetylated chromatin appears to be more “open”, and accessible to transcription factors and polymerases – HATs are implicated as co-activators of genes in chromatin, and HDACs are implicated as corepressors Matrix and scaffold The mitotic scaffold and nuclear matrix provide attachment sites for nuclear DNA Matrix Scaffold loops of duplex DNA Nucleus Mitotic chromosomes, with some DNA released In interphase chromosomes, at least some DNA is attached to a matrix Chromosome localization in interphase In interphase, chromosomes appear to be localized to a sub-region of the nucleus. Gene activation and location in the nucleus • Condensed chromatin tends to localize close to the centromeres – Pericentromeric heterochromatin • Movement of genes during activation and silencing – High resolution in situ hybridization – Active genes found away from pericentromeric heterochromatin – Silenced genes found associated with pericentromeric heterochromatin