ISI3
advertisement

The topology of point clouds:
galaxies, multiple sclerosis
lesions, and ‘bubbles’
Keith Worsley, Nicholas Chamandy, McGill
Jonathan Taylor, Stanford and Université de Montréal
Robert Adler, Technion
Philippe Schyns, Fraser Smith, Glasgow
Frédéric Gosselin, Université de Montréal
Arnaud Charil, Montreal Neurological Institute
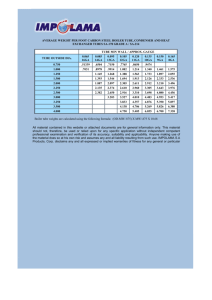
Astrophysics
Sloan Digital Sky
Survey,
release
6, Aug. ‘07
Sloan
Digital Skydata
Survey,
FWHM=19.8335
2000
Euler Characteristic (EC)
1500
1000
500
"Meat ball"
topology
"Bubble"
topology
0
-500
-1000
"Sponge"
topology
-1500
Observed
Expected
-2000
-5
-4
-3
-2
-1
0
1
Gaussian threshold
2
3
4
5
What is ‘bubbles’?
Nature (2005)
Subject is shown one of 40
faces chosen at random …
Happy
Sad
Fearful
Neutral
… but face is only revealed
through random ‘bubbles’
First trial: “Sad” expression
Sad
75 random
Smoothed by a
bubble centres Gaussian ‘bubble’
What the
subject sees
1
0.9
0.8
0.7
0.6
0.5
0.4
0.3
0.2
0.1
0
Subject is asked the expression:
Response:
“Neutral”
Incorrect
Your turn …
Trial 2
Subject response:
“Fearful”
CORRECT
Your turn …
Trial 3
Subject response:
“Happy”
INCORRECT
(Fearful)
Your turn …
Trial 4
Subject response:
“Happy”
CORRECT
Your turn …
Trial 5
Subject response:
“Fearful”
CORRECT
Your turn …
Trial 6
Subject response:
“Sad”
CORRECT
Your turn …
Trial 7
Subject response:
“Happy”
CORRECT
Your turn …
Trial 8
Subject response:
“Neutral”
CORRECT
Your turn …
Trial 9
Subject response:
“Happy”
CORRECT
Your turn …
Trial 3000
Subject response:
“Happy”
INCORRECT
(Fearful)
Bubbles analysis
1
E.g. Fearful (3000/4=750 trials):
+
2
+
3
+
Trial
4 + 5
+
6
+
7 + … + 750
1
= Sum
300
0.5
200
0
100
250
200
150
100
50
Correct
trials
Proportion of correct bubbles
=(sum correct bubbles)
/(sum all bubbles)
0.75
Thresholded at
proportion of
0.7
correct trials=0.68,
0.65
scaled to [0,1]
1
Use this
as a
0.5
bubble
mask
0
Results
Mask average face
Happy
Sad
Fearful
But are these features real or just noise?
Need statistics …
Neutral
Statistical analysis
Correlate bubbles with response (correct = 1, incorrect =
0), separately for each expression
Equivalent to 2-sample Z-statistic for correct vs. incorrect
bubbles, e.g. Fearful:
Trial 1
2
3
4
5
6
7 …
750
1
0.5
0
1
1
Response
0
1
Z~N(0,1)
statistic
4
2
0
-2
0
1
1 …
1
0.75
Very similar to the proportion of correct bubbles:
0.7
0.65
Results
Thresholded at Z=1.64 (P=0.05)
Happy
Average face
Sad
Fearful
Neutral
Z~N(0,1)
statistic
4.58
4.09
3.6
3.11
2.62
2.13
1.64
Multiple comparisons correction?
Need random field theory …
Euler Characteristic Heuristic
Euler characteristic (EC) = #blobs - #holes (in 2D)
Excursion set Xt = {s: Z(s) ≥ t}, e.g. for neutral face:
EC = 0
30
20
0
-7
-11
13
14
9
0
Heuristic:
At high thresholds t,
the holes disappear,
EC ~ 1 or 0,
E(EC) ~ P(max Z ≥ t).
Observed
Expected
10
EC(Xt)
1
0
-10
-20
-4
-3
-2
-1
0
1
Threshold, t
2
• Exact expression for
E(EC) for all thresholds,
• E(EC) ~ P(max Z ≥ t) is
3
4
extremely
accurate.
The»result
If Z(s) N(0; 1) ¡is an¢ isotropic Gaussian random ¯eld, s 2 <2 ,
with ¸2 I2£2 = V @Z ,
@s
µ
¶
P max Z(s) ¸ t ¼ E(EC(S \ fs : Z(s) ¸ tg))
s2S
Z 1
1
£
L (S)
= EC(S)
e¡z2 =2 dz
0
(2¼)1=2
t
L (S)
£ 1 e¡t2 =2
1
+
¸
Perimeter(S)
Lipschitz-Killing
1
2
2¼
curvatures of S
1
L (S)
¡t2 =2
2 Area(S) £
(=Resels(S)×c)
+
¸
te
2
(2¼)3=2
If Z(s) is white noise convolved
with an isotropic Gaussian
Z(s)
¯lter of Full Width at Half
Maximum
FWHM then
p
¸ = 4 log 2 :
FWHM
½0 (Z ¸ t)
½1 (Z ¸ t)
½2 (Z ¸ t)
EC densities
of Z above t
white noise
=
filter
*
FWHM
Results, corrected for search
Random field theory threshold: Z=3.92 (P=0.05)
Happy
Average face
Sad
Fearful
Neutral
Z~N(0,1)
statistic
4.58
4.47
4.36
4.25
4.14
4.03
3.92
3.82
3.80
3.81
3.80
Saddle-point approx (2007): Z=↑ (P=0.05)
Bonferroni: Z=4.87 (P=0.05) – nothing
Expected EC
Let s 2 S ½ <D .
Let Z(s) = (Z1 (s); : : : ; Zn (s)) be iid smooth Gaussian random ¯elds.
Let T (s) = f (Z(s)), e.g. the Â
¹ random ¯eld (n = 2):
T (s) =
max Z1 (s) cos µ + Z2 (s) sin µ:
0·µ ·¼=2
Let Xt = fs : T (s) ¸ tg be the the excursion set inside S.
Let Rt = fz : f (z) ¸ tg be the rejection region of T .
Then
X
D
\
L (S)½ (R ):
E(EC(S Xt )) =
d
d
t
d=0
Â
¹=
max Z1 cos µ + Z2 sin µ
0·µ·¼=2
Functions of Gaussian fields:
Z1~N(0,1)
Example:
3
Z2~N(0,1)
s2
2
1
0
-1
-2
-3
Excursion sets,
Xt = fs : Â
¹ ¸ tg
Search
Region,
S
Rt = fZ : Â
¹ ¸ tg
s1
Rejection regions,
Z2
Cone
2
alternative
4
0
2
Z1
Null
3
1
-2
-2
0
2
Threshold t
E(EC(S \ Xt )) =
Beautiful symmetry:
X
D
L (S)½ (R )
d
d
t
d=0
Lipschitz-Killing curvature Ld (S)
Steiner-Weyl Tube Formula (1930)
EC density ½d (Rt )
Taylor Gaussian Tube Formula (2003)
µ
¶
• Put a tube of radius r about
@Z the search region λS and rejection region Rt:
¸ = Sd
@s
Z2~N(0,1)
14
r
12
10
Rt
Tube(λS,r)
8
Tube(Rt,r)
r
λS
6
t-r
t
Z1~N(0,1)
4
2
2
4
6
8 10 12 14
• Find volume or probability, expand as a power series in r, pull off1coefficients:
jTube(¸S; r)j =
X
D
d=0
¼d
L
P(Tube(Rt ; r)) =
¡d (S)r d
D
¡(d=2 + 1)
X (2¼)d=2
d!
d=0
½d (Rt )rd
Lipschitz-Killing
curvature Ld (S)
of a triangle
r
Tube(λS,r)
λS
¸ = Sd
µ
@Z
@s
¶
Steiner-Weyl Volume of Tubes Formula (1930)
Area(Tube(¸S; r)) =
X
D
¼ d=2
L
¡d (S)r d
D
¡(d=2 + 1)
d=0
= L2 (S) + 2L1 (S)r + ¼ L0 (S)r2
= Area(¸S) + Perimeter(¸S)r + EC(¸S)¼r2
L (S) = EC(¸S)
0
L (S) = 1 Perimeter(¸S)
1
2
L (S) = Area(¸S)
2
Lipschitz-Killing curvatures are just “intrinisic volumes” or “Minkowski functionals”
in the (Riemannian) metric of the variance of the derivative of the process
Lipschitz-Killing curvature Ld (S) of any set
S
S
S
¸ = Sd
Edge length × λ
12
10
8
6
4
2
.
.. . .
.
. . .
.. . .
.. . .
.
. . . .
. . . .
. . . .
.. . .
. . .
... .
.
4
..
.
.
.
.
.
.
.
.
.
.
.
6
..
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
8
..
. .
. ...
. ..
. . .
. .
. . .....
. . .
. ....
..
..
10
µ
@Z
@s
¶
of triangles
L (Lipschitz-Killing
²) = 1, L (¡) curvature
L (N
=
1,
)=1
0
0
0
L (¡) = edge length, L (N) = 1 perimeter
1
2
L1 (N) = area
2
P Lcurvature
P L
Lipschitz-Killing
union
L
² ¡ Pof L
¡ of triangles
N
(S) = P² 0 ( )
¡ 0( ) +
P
L (S) =
L (¡) ¡
L (N)
¡
N 1
L1 (S) = P L 1(N)
2
N 2
0
N
0
( )
s2
Z~N(0,1)
Non-isotropic data?
µ
¸ = Sd
3
@Z
@s
¶
2
1
0.14
0.12
0
-1
-2
s1
0.1
0.08
0.06
-3
. . . .. .
12we warp
..
• Can
to isotropy? i.e. multiply edge lengths by λ?
... . .the
. . data
•
•
•
.
. . . . . . . .
10 .
. . . . . . . ...
. . no,
Globally
. . but
. . locally
. . . . yes, but we may need extra dimensions.
.
8 . . . . . . . . . .
. .
. . . . . . . Theorem:
Nash
Embedding
#dimensions ≤ D + D(D+1)/2; D=2: #dimensions
6 . . . . . . . . . .....
.. . . . . . . . .
. . . . Euclidean
....
4 idea:
. . . replace
Better
distance by the variogram:
... . . . . ...
. . . . .
d(s1, s2)2 = Var(Z(s1) - Z(s2)).
2
4
6
8
10
≤ 5.
Non-isotropic data
¸(s) = Sd
Z~N(0,1)
s2
3
µ
@Z
@s
¶
2
1
0.14
0.12
0
-1
-2
Edge length × λ(s)
12
10
8
6
4
2
..
.
.
.
. .
. .
.
.. .
.
.. .
.
.
.
.
.
.
.
. . .
.
. . .
.. .
.
.
. .
...
.
. .
.
.
.
.
.
.
.
.
.
.
.
4
6
..
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
8
.
..
. .
. ...
. ..
. . .
. .
. . .....
. . .
. ....
...
10
s1
0.1
0.08
0.06
-3
of triangles
L (Lipschitz-Killing
²) = 1, L (¡) curvature
L (N
=
1,
)=1
0
0
0
L (¡) = edge length, L (N) = 1 perimeter
1
2
L1 (N) = area
2
P Lcurvature
P L
Lipschitz-Killing
union
L
² ¡ Pof L
¡ of triangles
N
(S) = P² 0 ( )
¡ 0( ) +
P
L (S) =
L (¡) ¡
L (N)
¡
N 1
L1 (S) = P L 1(N)
2
N 2
0
N
0
( )
Estimating Lipschitz-Killing curvature Ld (S)
We need independent & identically distributed random fields
e.g. residuals from a linear model
Z1
Z2
Z3
Z4
Replace coordinates of
the triangles 2 <2 by
normalised residuals
Z 2<
n;
jjZjj
Z5
Z7
Z8
Z9 … Zn
of triangles
L (Lipschitz-Killing
²) = 1, L (¡) curvature
L (N
=
1,
)=1
0
0
0
L (¡) = edge length, L (N) = 1 perimeter
1
2
L1 (N) = area
2
P Lcurvature
P L
Lipschitz-Killing
union
L
² ¡ Pof L
¡ of triangles
N
(S) = P² 0 ( )
¡ 0( ) +
P
L (S) =
L (¡) ¡
L (N)
¡
N 1
L1 (S) = P L 1(N)
2
N 2
0
Z = (Z1 ; : : : ; Zn ):
Z6
N
0
( )
Scale space: smooth Z(s) with range of filter widths w
= continuous wavelet transform
adds an extra dimension to the random field: Z(s,w)
Scale space, no signal
w = FWHM (mm, on log scale)
34
8
6
4
2
0
-2
22.7
15.2
10.2
6.8
-60
-40
34
-20
0
20
One 15mm signal
40
60
8
6
4
2
0
-2
22.7
15.2
10.2
6.8
-60
-40
-20
0
s (mm)
20
40
60
15mm signal is best detected with a 15mm smoothing filter
Z(s,w)
Matched Filter Theorem (= Gauss-Markov Theorem):
“to best detect signal + white noise,
filter should match signal”
10mm and 23mm signals
w = FWHM (mm, on log scale)
34
8
6
4
2
0
-2
22.7
15.2
10.2
6.8
-60
-40
34
-20
0
20
Two 10mm signals 20mm apart
40
60
8
6
4
2
0
-2
22.7
15.2
10.2
6.8
-60
-40
-20
0
20
40
60
s (mm)
But if the signals are too close together they are
detected as a single signal half way between them
Z(s,w)
Scale space can even separate
two signals at the same location!
8mm and 150mm signals at the same location
10
5
w = FWHM (mm, on log scale)
0
-60
170
-40
-20
0
20
40
60
20
76
15
34
10
15.2
6.8
5
-60
-40
-20
0
s (mm)
20
40
60
Z(s,w)
Scale space Lipschitz-Killing curvatures
R
Suppose f is a kernel with f 2 = 1 and B is a Brownian sheet.
Then the scale space random ¯eld is
Z µ ¡ ¶
s h
Z(s; w) = w¡D=2 f
dB(h) » N(0; 1):
w
Lipschitz-Killing curvatures:
¡d
¡d
w
+
w
L (S £ [w ; w ]) = 1
2 L (S) +
d
1
2
d
2
b(D¡X
d+1)=2c
j=0
w¡d¡2j+1 ¡ w¡d¡2j+1
1
2
¡
d + 2j 1
¡
¡
¡
£ ·(1 2j)=2 ( 1)j (d + 2j 1)! L
¡1 (S);
¡
¡
d+2j
(1 2j)(4¼)j j!(d 1)!
where · =
R³
s0 @f (s)
@s
´
2
+ D f (s) ds. For a Gaussian kernel, · = D=2. Then
2
µ
¶ D+1
X
¸
¼
L (S £ [w ; w ])½ (R ):
P max Z(s) t
d
1
2
d
t
s2S
d=0
Rotation space:
Try all rotated elliptical filters
Unsmoothed data
Threshold
Z=5.25 (P=0.05)
Maximum filter
E(EC(S \ Xt )) =
Beautiful symmetry:
X
D
L (S)½ (R )
d
d
t
d=0
Lipschitz-Killing curvature Ld (S)
Steiner-Weyl Tube Formula (1930)
EC density ½d (Rt )
Taylor Gaussian Tube Formula (2003)
µ
¶
• Put a tube of radius r about
@Z the search region λS and rejection region Rt:
¸ = Sd
@s
Z2~N(0,1)
14
r
12
10
Rt
Tube(λS,r)
8
Tube(Rt,r)
r
λS
6
t-r
t
Z1~N(0,1)
4
2
2
4
6
8 10 12 14
• Find volume or probability, expand as a power series in r, pull off1coefficients:
jTube(¸S; r)j =
X
D
d=0
¼d
L
P(Tube(Rt ; r)) =
¡d (S)r d
D
¡(d=2 + 1)
X (2¼)d=2
d!
d=0
½d (Rt )rd
EC density ½d (Â
¹ ¸ t)
of the Â
¹ statistic
Z2~N(0,1)
Tube(Rt,r)
Â(s)
¹
=
r
max Z1 (s) cos µ + Z2 (s) sin µ
0·µ·¼=2
t-r
Taylor’s Gaussian Tube Formula
(2003)
1
P (Z1 ; Z2 2 Tube(Rt ; r)) =
X (2¼)d=2
d!
Rejection region
Rt
t
Z1~N(0,1)
½d (Â
¹ ¸ t)rd
d=0
(2¼)1=2 ½1 (Â
¹
¸ t)r + (2¼)½ (Â
¸ t)r2 =2 + ¢ ¢ ¢
= ½0 (Â
¹ ¸ t) +
¹
2
Z 1
=
(2¼)¡1=2 e¡z2 =2 dz + e¡(t¡r)2 =2 =4
t¡r
½0 (Â
¹ ¸ t) =
Z
t
1
(2¼)¡1=2 e¡z2 =2 dz + e¡t2 =2 =4
½1 (Â
¹ ¸ t) = (2¼)¡1 e¡t2 =2 + (2¼)¡1=2 e¡t2 =2 t=4
½ (Â
¹ ¸ t) = (2¼)¡3=2 e¡t2 =2 t + (2¼)¡1 e¡t2 =2 (t2 ¡ 1)=8
2
..
.
EC densities for some standard test statistics
Using Morse theory method (1981, 1995):
T, χ2, F (1994)
Scale space (1995, 2001)
Hotelling’s T2 (1999)
Correlation (1999)
Roy’s maximum root, maximum canonical correlation (2007)
Wilks’ Lambda (2007) (approximation only)
Using Gaussian Kinematic Formula:
T, χ2, F are now one line …
Likelihood ratio tests for cone alternatives (e.g chi-bar, beta-bar) and
nonnegative least-squares (2007)
…
Accuracy of the P-value approximation
If Z(s) » N(0; 1) ¡is an¢ isotropic Gaussian random ¯eld, s 2 <2 ,
with ¸2 I2£2 = V @Z ,
@s
µ
¶
P max Z(s) ¸ t ¼ E(EC(S \ fs : Z(s) ¸ tg))
s2S
Z 1
1
= EC(S) £
e¡z2 =2 dz
(2¼)1=2
t
1 ¡2
£
1
+ ¸ Perimeter(S)
e t =2
2
2¼
1
£
2
+ ¸ Area(S)
te¡t2 =2
(2¼)3=2
Z 1
1
= c0
e¡z2 =2 dz + (c1 + c2 t + ¢ ¢ ¢ + cD tD¡1 )e¡t2 =2
(2¼)1=2
t
¯ µ
¯
¶
¯
¯
¯P max Z(s) ¸ t ¡ E(EC(S \ fs : Z(s) ¸ tg))¯ = O(e¡®t2 =2 ); ® > 1:
¯
¯
s2S
The expected EC gives all the polynomial terms in the expansion for the P-value.
Bubbles task in fMRI scanner
Correlate bubbles with BOLD at every voxel:
Trial
1
2
3
4
5
6
7 …
3000
1
0.5
0
fMRI
10000
0
Calculate Z for each pair (bubble pixel, fMRI voxel)
a 5D “image” of Z statistics …
Thresholding? Cross correlation random field
Correlation between 2 fields at 2 different locations,
searchedµ
over all pairs of locations,
¶ one in S, one in T:
P
max C(s; t) ¸ c
s2S;t2T
=
¼ E(EC fs 2 S; t 2 T : C(s; t) ¸ cg)
dim(S)
X dim(T
X)
i=0
2n¡2¡h (i ¡ 1)!j!
¸
½ij (C c) =
¼h=2+1
L (S)L (T )½ (C ¸ c)
i
j
ij
j=0
b(hX
¡1)=2c
(¡1)k ch¡1¡2k (1 ¡ c2 )(n¡1¡h)=2+k
k=0
X
k X
k
l=0 m=0
¡( n¡i + l)¡( n¡j + m)
2
2
¡
¡
¡
¡
l!m!(k l m)!(n 1 h + l + m + k)!(i ¡ 1 ¡ k ¡ l + m)!(j ¡ k ¡ m + l)!
Cao & Worsley, Annals of Applied Probability (1999)
Bubbles data: P=0.05, n=3000, c=0.113, T=6.22
Discussion: modeling
The random response is Y=1 (correct) or 0 (incorrect), or Y=fMRI
The regressors are Xj=bubble mask at pixel j, j=1 … 240x380=91200 (!)
Logistic regression or ordinary regression:
logit(E(Y)) or E(Y) = b0+X1b1+…+X91200b91200
But there are only n=3000 observations (trials) …
Instead, since regressors are independent, fit them one at a time:
logit(E(Y)) or E(Y) = b0+Xjbj
However the regressors (bubbles) are random with a simple known distribution, so
turn the problem around and condition on Y:
E(Xj) = c0+Ycj
Equivalent to conditional logistic regression (Cox, 1962) which gives exact
inference for b1 conditional on sufficient statistics for b0
Cox also suggested using saddle-point approximations to improve accuracy of
inference …
Interactions? logit(E(Y)) or E(Y)=b0+X1b1+…+X91200b91200+X1X2b1,2+ …
MS lesions and cortical thickness
Idea: MS lesions interrupt neuronal signals, causing thinning in
down-stream cortex
Data: n = 425 mild MS patients
5.5
Average cortical thickness (mm)
5
4.5
4
3.5
3
2.5
Correlation = -0.568,
T = -14.20 (423 df)
2
1.5
0
10
20
30
40
50
Total lesion volume (cc)
60
70
80
MS lesions and cortical thickness at all pairs of
points
Dominated by total lesions and average cortical thickness, so remove these
effects as follows:
CT = cortical thickness, smoothed 20mm
ACT = average cortical thickness
LD = lesion density, smoothed 10mm
TLV = total lesion volume
Find partial correlation(LD, CT-ACT) removing TLV via linear model:
CT-ACT ~ 1 + TLV + LD
test for LD
Repeat for all voxels in 3D, nodes in 2D
~1 billion correlations, so thresholding essential!
Look for high negative correlations …
Threshold: P=0.05, c=0.300, T=6.48
Cluster extent rather than peak height
(Friston, 1994)
Choose a lower level, e.g. t=3.11 (P=0.001)
Find clusters i.e. connected components of excursion set
L (cluster)
Measure cluster
extent
by resels D
Z
D=1
extent
L (cluster) » c
D
t
®
k
Distribution of maximum cluster extent:
Bonferroni on N = #clusters ~ E(EC).
Peak
height
Distribution:
fit a quadratic to the
peak:
Y
s