American Cancer Society Study
advertisement
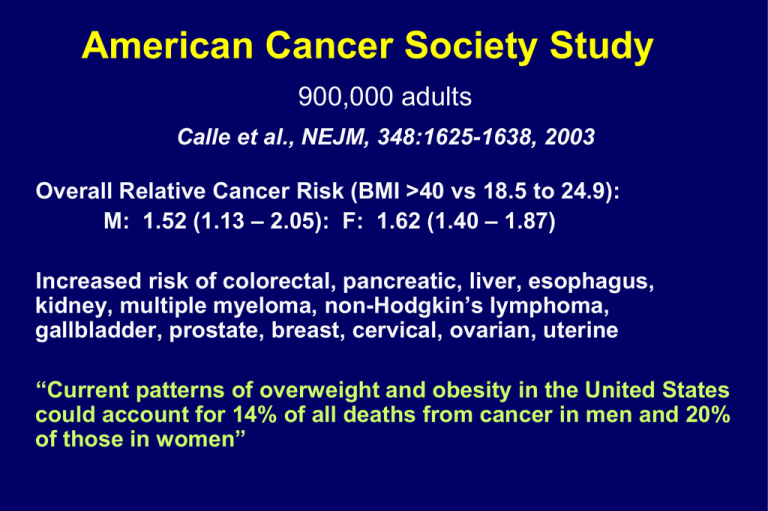
American Cancer Society Study 900,000 adults Calle et al., NEJM, 348:1625-1638, 2003 Overall Relative Cancer Risk (BMI >40 vs 18.5 to 24.9): M: 1.52 (1.13 – 2.05): F: 1.62 (1.40 – 1.87) Increased risk of colorectal, pancreatic, liver, esophagus, kidney, multiple myeloma, non-Hodgkin’s lymphoma, gallbladder, prostate, breast, cervical, ovarian, uterine “Current patterns of overweight and obesity in the United States could account for 14% of all deaths from cancer in men and 20% of those in women” Guidelines for Healthy Weight Nurses’ Health Study: Willett et al., NEJM, 341:427-434, 1999 BMI of 26 vs 21 Coronary Heart Disease: Hypertension: Type II Diabetes: 2x increase 2-3x increase 8x increase Weight Change of 15 kg Coronary Heart Disease: Hypertension: Type II Diabetes: 2x increase 2-3x increase 6x increase Caloric Restriction (CR) CR is an experimental paradigm in which the dietary/caloric intake of a group of animals is reduced relative to that eaten by ad libitum fed controls Caloric restriction is the most potent, most robust, and most reproducible known means of reducing morbidity and mortality in mammals Survival Data, 1987 Cohort, Casein Diet Survival Rate 100 80 AL DR 60 40 20 0 0 200 400 600 800 Days 1000 1200 1400 DR Reduces Morbidity: Breast Cancer • Delays tumor onset (initiation and promotion) • Slows progression • Can modulate oncogene penetrance – v-Ha-ras tumors decreased 67% • (Fernandes et al., PNAS 92:6494-6498, 1995) • Can prevent carcinogen-induced tumors – 7,12-demethyl-benz(a)anthracene (60% AL, 0% DR) • Kritchevsky et al. Cancer Res 44, 3174-3177, 1984 – Even with high fat diet, tumor yield, size, burden down 93-98% • Klurfied et al. Cancer Res 47, 2759-2762, 1987 CR “Beneficial” Effects • Lower oxidative stress – “Better” redox balance • “Improved” glucose metabolism – Increased insulin sensitivity – Reduced blood glucose – Reduced diabetes risk • Reduced inflammation How do we study complex biological/clinical problems? How do we address such questions in humans, where our ability to manipulate and analyze the system is limited? High Throughput and/or Data Density Studies • • • • Genomics/SNPs mRNA expression arrays Proteomics Small metabolites Metabolomics: The –omics face of biochemistry Measurement of changes in populations of low molecular weight metabolites under a given set of conditions Fiehn GENOME TRANSCRIPTOME PROTEOME DISEASE STATE METABOLOME ENVIRONMENT HEALTH STATE GENOME TRANSCRIPTOME PROTEOME DISEASE STATE METABOLOME ENVIRONMENT HEALTH STATE Sample Analysis Sample Collection Database Curation Response (µA) 0.80 0.60 0.40 0.20 0.00 0.0 20.0 40.0 60.0 80.0 Retention time (minutes) 1 100.0 Objectively Defining Class Identity 3 SD 2 SD Actual Mechanistic Insight Drug Development Toxicology Classification Prediction Functional genomics Sub-threshold studies Others AL8 AL7 AL5 AL1 AL4 AL3 AL2 AL6 DR8 DR6 DR5 DR7 DR1 DR4 DR2 DR3 1.0 Computational Modeling of Metabolic Serotypes Observed Values vs. Predicted Values 0.8 0.6 0.4 0.2 0.0 2 SD Predicted Modeling Metabolic Interactions Following Biochemical Pathways Bioinformatics What we measure -- biochemically Metabolites – small molecules Pathways (eg, purine catabolites) Interactive pathways (eg, amino acid metabolism) Compound classes (eg, lipids) Conceptually linked systems eg antioxidants, redox damage products What we measure -- conceptually Biochemical constituents Excretion products Precursor – product Balances (eg, redox systems) “collection depots” Flux Snapshot view of biochemistry Integrated signal from genome and environment Short and long term status Temporal image Sub-threshold changes (eg (toxicology, nutrition) Metabolomics – Some Advantages Sensitivity “silent phenotypes”/sub-threshold effects Discovery Knowledge base (ie, metabolic pathways) Limited repertoire – simplifies possibilities (2500 non-lipid endogenous metabolites??) Metabolome integrates signal Nature and Nurture -- genome and environment Measurement of system status/defects Metabolome has the fastest response time Metabolomics – Some Disadvantages Too Sensitive? cohort effects, site effects, time effects sample handling individual metabolites responsive to multiple factors genes, environment, health status, location experiment design must account for all factors controlled or fuzzy, multiple sources Practical Set-up costs Possible need for multiple platforms (NMR, MS, HPLC) early industry dominance – lots of propriety data incompatible data standards Metabolomics Technology = Metabolomics Platform Biology Analytical Chemistry Data Analysis Sample Analysis Sample Collection Database Curation Response (µA) 0.80 0.60 Analytical 0.40 0.20 0.00 0.0 20.0 40.0 60.0 80.0 Retention time (minutes) 1 100.0 Objectively Defining Class Identity 3 SD 2 SD Actual Mechanistic Insight Drug Development Toxicology Classification Prediction Functional genomics Sub-threshold studies Others AL8 AL7 AL5 AL1 AL4 AL3 AL2 AL6 DR8 DR6 DR5 DR7 DR1 DR4 DR2 DR3 1.0 Computational Modeling of Metabolic Serotypes Observed Values vs. Predicted Values 0.8 0.6 0.4 0.2 0.0 2 SD Predicted Modeling Metabolic Interactions Following Biochemical Pathways Bioinformatics Sample Analysis Sample Collection Database Curation Response (µA) 0.80 0.60 0.40 0.20 0.00 0.0 20.0 40.0 60.0 80.0 Retention time (minutes) 1 Data Analysis 100.0 Objectively Defining Class Identity 3 SD 2 SD Actual Mechanistic Insight Drug Development Toxicology Classification Prediction Functional genomics Sub-threshold studies Others AL8 AL7 AL5 AL1 AL4 AL3 AL2 AL6 DR8 DR6 DR5 DR7 DR1 DR4 DR2 DR3 1.0 Computational Modeling of Metabolic Serotypes Observed Values vs. Predicted Values 0.8 0.6 0.4 0.2 0.0 2 SD Predicted Modeling Metabolic Interactions Following Biochemical Pathways Bioinformatics Sample Analysis Sample Collection Database Curation Response (µA) 0.80 0.60 0.40 0.20 0.00 20.0 40.0 60.0 80.0 Retention time (minutes) Biology Mechanistic Insight Drug Development Toxicology Classification Prediction Functional genomics Sub-threshold studies Others 1 100.0 Objectively Defining Class Identity AL8 AL7 AL5 AL1 AL4 AL3 AL2 AL6 DR8 DR6 DR5 DR7 DR1 DR4 DR2 DR3 1.0 Computational Modeling of Metabolic Serotypes Observed Values vs. Predicted Values 3 SD 2 SD Actual 0.0 0.8 0.6 0.4 0.2 0.0 2 SD Predicted Modeling Metabolic Interactions Following Biochemical Pathways Bioinformatics Caloric intake as a case study High points only Ignoring details, other studies, etc Survival Data, 1987 Cohort, Casein Diet Survival Rate 100 80 AL DR 60 40 20 0 0 200 400 600 800 Days 1000 1200 1400 Hypothesis: Long-term, low-calorie diets induce changes in metabolism that persist throughout the lifespan Predictions • CR alters the sera “metabolome” • There exists a “CR Serotype” • …Part of “CR serotype” reflects beneficial physiological status --- ie, serotype defines health without reference to disease… Goals 1) 2) Insights into the mechanism of CR Recognize CR in other organisms (e.g., non-human primates) 3) Biochemically determine the effective, longterm caloric intake of an individual (e.g., for epidemiological studies) 4) Identify predictive markers of disease (e.g., to intervene/prevent/focus resources; focus on diseases where intervention is possible) Experimental Design Model: F344 x BN F1 Rat Overall Design: AL/CR, male/female, 5 different ages Different extents and duration of diets Total experiment ~36 groups, 82 cohorts. Approach: HPLC separations with coulometric array detection (LC/LC-MS for plasma proteomics) Multilayer statistical and data analysis Analytical Stability Biologic Variability Analytical vs Biological Variation 0.75 r ² = 0.75 0.36 0.24 0.12 0.00 0.00 AvA 0.45 0.45 0.30 0.30 0.15 0.15 0.12 0.24 0.36 0.48 0.00 0.00 0.60 0.15 0.30 0.45 0.60 0.00 0.00 0.75 0.15 0.30 r ² = 0.31 2.00 0.45 0.60 AL vs DR ANALCV2 ANALCV1 1.50 r ² = 0.36 0.60 0.60 ANALCVT ANALCVT 0.48 0.75 r ² = 0.55 ANALCV2 0.60 r ² = 0.25 1.50 r ² = 0.31 1.75 0.75 ANALCV1 2.00 1.50 0.50 1.25 FALCV 0.75 1.00 0.75 FDRCV 1.00 1.00 FDRCV MDRCV 1.50 0.75 1.00 0.75 0.50 0.25 0.25 0.25 0.25 0.50 0.75 1.00 1.25 0.00 0.00 1.50 0.25 0.25 0.50 0.75 1.00 1.25 1.50 1.75 0.00 0.00 2.00 0.25 0.50 0.75 FALCV MALCV 1.00 1.25 0.00 0.00 1.50 0.25 0.50 0.75 1.00 1.25 1.50 1.25 r ² = 0.01 0.75 0.50 2.00 2.00 r ² = 0.00 r ² = 0.01 1.00 1.50 ANALCV1 ANALCV1 1.00 1.75 1.75 1.25 1.00 0.75 0.50 ANALCV1 r ² = 0.01 1.25 1.50 MDRCV MALCV Biological vs Analytical 1.50 ANALCV1 1.25 0.50 0.50 0.00 0.00 r ² = 0.09 1.75 1.25 1.25 0.75 0.50 1.25 1.00 0.75 0.50 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.50 0.75 MALCV 1.00 1.25 1.50 0.00 0.00 0.25 0.50 0.75 1.00 MDRCV 1.25 1.50 0.00 0.00 0.25 0.50 0.75 FALCV 1.00 1.25 0.00 0.00 0.25 0.50 0.75 1.00 1.25 1.50 1.75 2.00 FDRCV In Rats: Biological variability 5 fold greater than analytical variability Analytical variability does not influence biological variability Primary Data Analysis • Multivariate analyses are relatively noise-resistant • Minimize loss of informative metabolites • Reduce false negatives (Type II errors) • Increase false positives (Type I errors) Does Serotype Encode Sufficient Information to Identify Diet Group? Data Exploration and Classification Analysis • Hierarchical Cluster Analysis (HCA) – Identifies natural groups in data • Principal Component Analysis (PCA) – Finds linear combinations of original variables that account for maximal variation Model Feature Selection Biological Model 0.80 AL DR Response (µA) Survival Rate 80 60 40 0.60 0.40 0.20 0.00 20 0.0 0 T-tests, p<0.2 ?! 1075 analytically detectable peaks sensitivity ~300 pA = ~10 fmole/125 l sera 100 20.0 40.0 60.0 80.0 100.0 16 15 14 13 12 11 10 9 8 7 6 5 4 3 2 1 350 300 250 200 150 100 50 Retention time (minutes) 0 200 400 600 800 1000 1200 0 1400 Days HCA Proof of Principle HCA Distinguishes Female AL and DR Rats Single Autoscale Range Scale Linkage Method Dependent Categorization Accuracy AL8 AL7 AL5 AL1 AL4 AL3 AL2 AL6 DR8 DR6 DR5 DR7 DR1 DR4 DR2 DR3 AL8 AL5 AL7 AL1 AL4 AL3 AL6 AL2 DR8 DR7 DR1 DR4 DR2 DR3 DR6 DR5 Complete AL DR AL DR 1.0 1.0 0.8 0.8 0.6 0.4 0.6 0.4 100% AL8 AL7 AL1 AL5 AL6 AL4 AL3 AL2 0.2 0.0 DR8 DR7 DR1 DR6 DR5 DR4 DR2 DR3 AL8 AL5 AL7 AL1 AL6 AL4 AL3 AL2 0.2 0.0 DR8 DR7 DR1 DR6 DR5 DR4 DR2 DR3 AL DR AL DR 1.0 0.8 1.0 0.8 0.6 0.6 100% 0.4 0.4 AL8 AL7 AL1 AL5 AL6 AL4 AL3 AL2 DR8 DR7 DR1 DR6 DR5 DR4 DR2 DR3 0.2 AL8 AL5 AL7 AL1 AL6 AL4 AL3 AL2 0.2 0.0 DR8 DR7 DR1 DR6 DR5 DR4 DR2 DR3 AL DR AL DR 1.0 1.0 0.8 0.8 0.6 0.6 100% 63 variables (confirmed), NonNon-independent samples PCA Distinguishes AL and DR Female Rats Preprocessing Dependent Categorization Accuracy Centroid 0.4 0.4 PCA Autoscale Range Scale 100% Factor1 Factor1 0.2 Factor2 0.0 DR 100% 0.2 Factor3 AL DR Factor2 Factor3 0.0 63 variables (confirmed), NonNon-independent samples AL HCA Validation PCA Distinguishes AL and DR Female Rats Complete 1.0 0.8 0.6 0.4 0.2 PCA 0.0 Factor3 AL11 DR16 DR15 DR13 DR14 DR12 DR11 DR9 DR10 AL16 AL15 AL12 AL10 AL13 DR AL DR Factor2 AL Factor1 AL9 AL14 63 variables (confirmed), independent female Cohort #2 94% Accuracy HCA Simplify Model PCA PCA Distinguishes AL/DR Using Either Dataset Removed variables can contribute to AL-DR separations 63 Variables Autoscale, complete 63 Variables 1.0 0.8 0.6 0.4 36 Variables 0.2 0.0 1.0 DR21 DR21 AL21 AL21 AL17 AL18 AL20 AL AL19 AL22 AL19 AL22 AL18 DR19 AL20 DR18 DR DR17 91% Accuracy 0.6 0.4 AL Factor2 0.2 0.0 Factor3 AL AL Factor1 Factor1 Factor2 DR19 AL17 DR20 0.8 36 Variables DR20 DR18 DR17 DR 63% Accuracy 63/(37/36) variables (confirmed), Independent female Cohort #3 DR DR Factor3 37 variables (confirmed), Independent female Cohort #3 Status Proof of principle accuracy: HCA (100%) PCA (100%) Validation Accuracy: HCA (94%) PCA (100%) - subjective rotation Simplification – HCA (Fails) PCA (100% Accuracy) Use larger models? Test components vs distance “Expert Systems/Supervised Analysis” KNN – – – – k-nearest neighbor analysis Supervised HCA (HCA is KNN with K=1) Distance-based metric Strength is with small (training) datasets SIMCA – – – – Soft Independent Modeling of Class Analogy Supervised PCA Component-based metric Strength is modeling flexibility (eg, group-specific interactions) In our DR sera metabolomics data – components greatly outperform distance-based algorithms In OUR DR SERA METABOLOMICS data – components greatly outperform distance-based algorithms Profiles are cohort specific Cohort Separations male samples modeled with male/female data set 4 2 0 -2 -4 -6 6 4 2 0 -2 8 6 -4 4 2 0 -2 -8-6 -4 AMAL AMDR BMAL BMDR CMAL CMDR Cohort Effects 2 Cohort Separations 4 10 8 6 4 2 0 -2 -4 -6 -8 6 4 t[1 ] 4 -2 -4 -6 -8 8 -2 4 -6 -8 -10 -6 2 0 -2 t[2] -10 -4 winsorize (3SD) -4 6 -8 0 log winsorize (2SD) winsorize (3SD) 2 0 -2 10 -6 2 log winsorize (2SD) 6 0 t[1 ] t[3] t[3] AMAL AMDR BMAL BMDR CMAL CMDR -4 log + win (2SD) -8 -6 -8 -12 log + win (2SD) -10 -10 log + win (3SD) log + win (3SD) 0.0 (d) log + win (3SD) (c) winsorize (3SD) 0.2 0.4 0.6 0.8 1.0 0.0 (c) Pareto scaling (females) 0.2 0.4 0.6 0.8 1.0 (d) Pareto scaling (males) none none log 8 log 6 6 4 8 2 0 -2 -4 -6 -8 8 t[3] 4 -2 0 -2 8 -4 6 4 6 0 2 0 -8 -2 -4 -6 -8 -4 -6 -8 -8 -10 -12 log + win (3SD) -10 log + win (3SD) 0.0 (f) log + win (2SD) t[1 (e) winsorize (2SD) log + win (2SD) -6 -2 t[2] -10 winsorize (3SD) log + win (2SD) -4 4 -6 2 t[2] winsorize (3SD) 2 -4 -6 -8 t[1 ] -2 10 winsorize (2SD) 6 0 2 0 winsorize (2SD) 8 2 6 4 t[1 ] 4 t[3] t[3] 6 4 2 0 -2 8 6 -4 4 2 -6 0 -2 -8 -4 t[2] (b) no scaling (males) none 8 2 8 6 4 2 0 -2 -4 8 t[2] 0 -2 -4 -6 (a) no scaling (females) 6 10 2 PLS-DA RX 2 RY 2 Q (cum) (b) log none male samples modeled with male/female data set 4 AMAL AMDR BMAL BMDR CMAL CMDR (a) none 0.2 0.4 0.6 0.8 0.0 1.0 ] (e) UV scaling (females) none 6 8 0.4 0.6 0.8 1.0 0.8 1.0 none log log winsorize (2SD) winsorize (2SD) winsorize (3SD) winsorize (3SD) log + win (2SD) log + win (2SD) 4 6 t[3] 4 2 -4 6 4 2 t[1 ] -2 8 -2 -4 -8 -6 -8 -10 4 -2 -6 0 t[2] 6 0 -4 -6 -8 10 0 10 8 2 6 2 0 -2 -4 -6 -8 2 0 -2 8 6 4 t[1 ] 4 t[3] 0.2 (f) UV scaling (males) -4 2 0 -6 -2 t[2] -4 -6 -8 -8 -10 -10 -12 log + win (3SD) log + win (3SD) -10 0.0 0.2 0.4 0.6 0.8 0.0 1.0 0.2 0.4 0.6 PLS-DA 8 t[2] 0 .4 0 -6 -12 -10 -8 - 0 .2 0 Comp[3] 0 -4 0 .0 0 Comp[2] 2 -2 0 .2 0 Comp[1] 54.808 0 .3 0 0.00 0.10 0.20 0.30 0.40 0.50 0.60 0.70 0.80 0.90 1.00 -6 -4 -2 0 2 t[1] 4 6 8 AL 0 .2 0 0.80 7.642 0.60 0.40 0 .0 0 -0 .1 0 -0 .2 0 0.20 CR 10 12 -0 . 1 0 0.00 0 .0 0 0 .1 0 w*c[1] Predicted AL probability 46.142 45.417 20.167.642 40.825 34.783 34.992 23.5 13.525 55.658 75.483 30.242 61.092 70.017 34.9 9.817 52.858 31.617 45.308 9.44216.933 8.183 21.117 13.467 47.442 41.642 75.525 62.942 68.575 68.6 11.45 30.133 54.058 56.117 48.808 14.975 24.483 6.408 49.6 68.7 40.4 82.225 27.892 92.792 22.192 17.475 62.492 77.917 40.292 28.608 12.608 65.408 28.858 63.358 15.475 48.533 76.2 62.65 100.242 9.4593.892 46.725 46.575 43.033 29.317 84.808 22.467 5.067 77.442 25.492 44.325 16.925 26.392 54.283 5.042 40.367 35.5 27.142 69.05 37.358 44.225 39.417 47.9675.758 72.342 52.108 58.167 47.192 11.458 14.033 73.075 0 .1 0 w*c[2] 4 0 .6 0 p<0.001 1.00 6 0 .8 0 Actual AL probability 1 .0 0 0 .9 0 0 .8 0 0 .7 0 0 .6 0 0 .5 0 0 .4 0 0 .3 0 0 .2 0 0 .1 0 0 .0 0 0 .2 0 110 100 N = 90 Each group p<0.05 vs others 77.917 L 90 K 80 70 60 r ² = 0.877; r ² = 0.994 (m eans) 50 40 40 50 60 70 80 90 100 110 16.92527.142 120 41.642 22.192 INTAKE 120 w*c[2] Predicted Degree Restriction Markers “Predict” Caloric Intake with High Quantitative Accuracy -- Proof of Concept -- 34.783 70.017 21.117 8.183 77.867 68.7 84.808 44.325 13.525 6.408 26.392 56.117 54.808 52.85815.475 68.575 68.6 75.483 82.225 28.608 48.533 81.7 17.475 47.967 30.133 54.058 65.408 28.858 45.417 62.942 35.5 92.792 77.442 29.317 16.933 44.225 40.292 62.492 30.108 100.242 30.242 7.642 58.167 5.067 46.142 13.467 55.658 37.358 34.483 67.642 72.933 40.4 43.033 61.092 62.65 5.758 9.45 62.442 47.442 47.192 23.5 54.283 34.992 9.442 14.975 52.108 63.358 34.9 45.308 27.892 73.075 49.6 76.2 24.483 5.042 11.308 75.525 48.808 11.45 11.458 39.417 Actual Degree of Restriction w*c[1] 25.492 O-PLS models built for better testing female basic model.M1 (OPLS) t[Comp. 1]/t[Comp. 2] Colored according to value in variable female basic model(Group Y new) Series (Variable Group Y new) 1 - 1.5 1.5 - 2 7 6 5 4 3 2 t[2]O 1 0 -1 -2 -3 -4 -5 -6 -7 -8 -8 -7 -6 -5 -4 -3 -2 -1 0 1 2 3 4 5 6 7 t[1]P R2X[1] = 0.105822 R2X[2] = 0.162921 Ellipse: Hotelling T2 (0.95) SIMCA-P 11 - 3/22/2006 2:34:11 PM 8 Iteratively improve models – focus on analytical robustness Then test one model… O-PLS Prediction Score (A.U.) Validation: Across Lifespan 6 8 4 6 2 4 2 0 0 -2 -2 -4 -4 -6 6 12 18 24 30 AL 6 12 18 24 30 [Months of Age] AGE CR -6 0 2 Validation: Duration 8 6 6 4 4 r²=0 2 2 0 0 -2 -2 -4 -4 4 30 CR -6 -6 0 2 4 6 8 10 12 14 16 18 Weeks on CR Diet 0 Validation: Extent 6 r ² = 0.645 4 2 0 -2 -4 -6 16 18 0 10 20 30 Percent Restriction 40 OPLS S 0 -2 Validation: Extent -4 0 10 20 30 40 50 Percent Restriction 6 4 Only including 8 week CR No AL group 8 r ² = 0.338 6 Includes 12 and 18 AL and CR r ² = 0.636 r ²0.636 OPLS Score AU OPLS Score AU 4 2 0 2 0 -2 -4 -2 -6 -8 -4 0 10 20 30 Percent Restriction 8 6 Includes 12 and 18 AL and CR r ² = 0.636 r ²0.636 40 50 0 10 20 Percent Restriction 30 40 Experimental Design Analytical Issues AL vs DR Biological Issues Analytical vs Biological Variation 0.75 r ² = 0.75 0.36 0.24 0.12 0.00 0.00 AvA 0.45 0.45 0.30 0.30 0.15 0.15 0.12 0.24 0.36 0.48 0.00 0.00 0.60 0.15 0.30 0.45 0.60 0.00 0.00 0.75 0.15 0.30 r ² = 0.31 2.00 0.45 0.60 AL vs DR ANALCV2 ANALCV1 1.50 r ² = 0.36 0.60 0.60 ANALCVT ANALCVT 0.48 0.75 r ² = 0.55 ANALCV2 0.60 r ² = 0.25 1.50 r ² = 0.31 1.75 0.75 ANALCV1 2.00 1.50 0.50 1.25 FALCV 0.75 1.00 0.75 FDRCV 1.00 1.00 FDRCV MDRCV 1.50 0.75 1.00 0.75 0.50 0.25 0.25 0.25 0.25 0.50 0.75 1.00 1.25 0.00 0.00 1.50 0.25 0.25 0.50 0.75 1.00 1.25 1.50 1.75 0.00 0.00 2.00 0.25 0.50 0.75 FALCV MALCV 1.00 1.25 0.00 0.00 1.50 0.25 0.50 0.75 1.00 1.25 1.50 1.25 r ² = 0.01 0.75 0.50 2.00 2.00 r ² = 0.00 r ² = 0.01 1.00 1.50 ANALCV1 ANALCV1 1.00 1.75 1.75 1.25 1.00 0.75 0.50 ANALCV1 r ² = 0.01 1.25 1.50 MDRCV MALCV Biological vs Analytical 1.50 ANALCV1 1.25 0.50 0.50 0.00 0.00 r ² = 0.09 1.75 1.25 1.25 0.75 0.50 1.25 1.00 0.75 0.50 0.25 0.25 0.25 0.25 0.00 0.00 0.25 0.50 0.75 MALCV 1.00 1.25 1.50 0.00 0.00 0.25 0.50 0.75 1.00 MDRCV 1.25 1.50 0.00 0.00 0.25 0.50 0.75 FALCV 1.00 1.25 0.00 0.00 0.25 0.50 0.75 1.00 1.25 1.50 1.75 2.00 FDRCV In Rats: Biological variability 5 fold greater than analytical variability Analytical variability does not influence biological variability Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Human Studies: Profile = … • Recent Food (ie, Fast) –Effect size ~ 0.2*StDev • BMI • Food intake • Physiology Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology BMI Profile Score Does Not Reflect BMI 45 45 40 40 35 35 30 30 25 25 20 20 15 15 -8 -6 -4 -2 0 2 4 Profile Score (Individual) 6 8 -8 -6 -4 -2 0 2 4 Profile Score (3 point mean) 6 Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Human Studies: FFQ vs score (individuals) y = 0.001x - 1.9244 R² = 0.033 8 6 4 2 0 0 -2 -4 -6 -8 500 1000 1500 2000 2500 3000 Extreme FFQ Quintiles I • No obvious association with individual FFQs, but… • FFQs are known to be weakly predictive of individual caloric intake • But extreme quintiles on the FFQ should differ – or we would likely never see anything in epidemiology Extreme FFQ Quintiles II 0.60 0.40 0.20 y = 0.0011x - 2.0509 R² = 0.834 0.00 -0.20 0 -0.40 1000 2000 3000 Individual values in extreme quintiles differ, p<0.005 -0.60 -0.80 FFQ Quintiles “predict” score Extreme FFQ Quintiles II 0.60 0.40 0.20 y = 772.21x + 1881.5 R² = 0.8336 y = 0.0011x - 2.0509 R² = 0.834 2500 2000 0.00 -0.20 0 3000 1500 1000 2000 3000 1000 -0.40 500 -0.60 0 -0.80 -1.00 -0.50 FFQ Quintiles “predict” score Score “predicts” FFQ quintile 0.00 0.50 Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Testing Physiology • Hypothesis: Profile score will differ/predict future risk of diseases reduced by CR, eg, breast cancer • Proposed 1000 cases/paired controls for breast cancer – Nested within Nurses’ Health Study – Samples taken 2-10 years before onset • Initial funding, 2 years, 750 case control pairs • Today, 210 case:control pairs (first blind break) Case-Controls - Design Using the raw RAT models, no corrections, no optimization except for those that addess analytical issues in humans All observations that served as both cases and controls (due to conversion) were excluded for this analysis 210 total case/control pairs scored The original model currently the best, so I'll present those numbers...others are close Case-Controls - Data These slides not cleared for posting at cohort study level, contact me at bkristal@partners.org if you have questions Human Studies: Profile = … • • • • Recent Food (ie, Fast) BMI Food intake Physiology Summary Created and validated a working model of the CR serotype in both male and female rats Profiles distinguish diet in blinded studies Markers pass analytical tests in human plasma Rat profiles pass key tests in human case/control studies Other studies ongoing, lipids, macronutrient shifts The metabolomic markers and profiles identified appear analytically and biologically suitable for studies in defined human populations such as national clinical trials and epidemiological cohorts Data suggests that the protective phenotype exemplified in the CR rat is broadly conserved across species Data suggests that we have the “raw material” needed to build marker profiles that can be tailored to provide a continuous ruler for objective measurement of factors such as caloric intake in epidemiological and clinical studies Data suggests that we have the “raw material” to begin to address personalized disease risk from the nutritional/environmental side Long-Range Disease Risk Prediction – Algorithmic Information Fusion in a Life and Death Environment Opportunities Genetics Demographics Questionnaires Clinical data Metabolomics, proteomics, lipidomics = GxE Long-Range Disease Risk Prediction – Algorithmic Information Fusion in a Life and Death Environment Complications Genetics…low effect size, low proportion explained, low penetrance, multi-gene effects, sequence vs SNP, Epigenome vs sequence, tissue specificity Demographics…definition and measurement problems Questionnaires…people lie, people are biased liars, questions are *hard* and often population specific Clinical data… limited, costly, by the time you have it may be too late, thresholds, different data structure/distributions Metabolomics… proteomics, lipidomics = GxE, very complex interactions, time effects, diet effects Long-Range Disease Risk Prediction – Algorithmic Information Fusion in a Life and Death Environment Complications Are we limited to decision level analysis? Long-Range Disease Risk Prediction – Algorithmic Information Fusion in a Life and Death Environment Complications Are we limited to decision level analysis? No… Biology-based fusion works Long-Range Disease Risk Prediction – Algorithmic Information Fusion in a Life and Death Environment Step 1 What is Success? What is Failure? How do we balance Success or Failure What does it mean to optimize? How do we assess? Long-Range Disease Risk Prediction – Algorithmic Information Fusion in a Life and Death Environment Step 1 What is Success? What is Failure? How do we balance Success or Failure What does it mean to optimize? How do we assess? Long-Range Disease Risk Prediction Algorithmic Information Fusion in a Life and Death Environment Long-Range Disease Risk Prediction Algorithmic Information Fusion in a Life and Death Environment Long-Range Disease Risk Prediction Algorithmic Information Fusion in a Life and Death Environment Long-Range Disease Risk Prediction Algorithmic Information Fusion in a Life and Death Environment Acknowledgements • Brigham and Women’s Hospital and Burke/Cornell – Yevgeniya Shurubor, Honglian Shi, Diane Sheldon, Sophie Guo, Susan (Schiavo) Bird, Rose Gathungu – Ugo Paolucci, Ruiwen Zhou, Vasant Marur, Neil Russell, Matt Sniatynski • Outside Collaborators – Walter Willett, Sue Hankinson, Frank Hu, Paul Vouros – Wayne Matson, Karen Vigneau-Callahan, Paul Milbury – Tom Vogl, Frank Hsu, Christina Schweikert • Funding – NIH (NIA; NCI; NIEHS) – Winifred Masterson Burke Relief Foundation – Brigham and Women’s Hospital, and Dept of Neurosurgery Acknowledgments Bioinformatics Analytical Work Animal Studies and Mitochondria Research