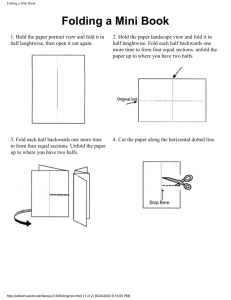
Recursive Domains in Proteins
advertisement

Recursive domains in proteins Teresa Przytycka NCBI, NIH Joint work with G.Rose & Raj Srinivasan; JHU Domain: “Polypeptide chain (or a part of it) that can independently fold into stable tertiary structure” (Baranden & Tooze; Introduction to Protein Structure) Two-domain protein. The 3D structure of a protein domain can be described as a compact arrangement of secondary structures Alpha helix Beta strand These arrangements are far from random: There are not so many of them : PDB contains about 17000 structures and less than 1000 different folds. Proportion of "new folds" (light blue) and "old folds" (orange) for a given year. (fold = fold domain) Possible sources of restricted number of folds: • Evolutionary history. – Given enough time would domains look “more random”? • Existence of general restrictions/rules which render some (compact) arrangements of secondary structures non-feasible. – Can real protein domains be seen as sentences in a language, which can be generated by an underlying grammar? Can protein domains be described using a set of folding rules? We restrict our attention to all beta domains: • they admit variety of topologies • they are difficult to predict from sequence Understanding b-folds • Patterns in b-sheets – Richardson 1977 • folding rules for b-sheets – Zhang and Kim 2000 • Hydrogen bonding pattern • Polypeptide chain seems to avoid “complications” Parallel anti-parallel mixed • Properties of b-sandwiches – Woolfson D. N., Evans P. A., Hutchinson E. G., and Thornton J. M. 1993 “forbidden” crossed conformation Expectations for good folding rules • We need to look at fold properties that occur in non-homologous proteins. • Preferably: The provide a model for the folding process. Super-secondary structures as precursors of folding rules • Super-secondary structure – frequently occurring arrangements of a small number of secondary structures • The occurrences of super-secondary structures in unrelated families supports possibility of their independent formation. Example 1: Hairpin b-b-b-unit Example 2: Greek key and suggested folding pathway for it Folding pathway for Greek key proposed by Ptitsyn. Pattern from a Greek vase Two level of folding rules: • Primitive folding rules – based on super secondary structures • Closure operation – allows for hierarchical application of the primitive rules supersecondary structures -primitive folding rules hairpin Hairpin rule Bridge Greek key Direct wind Indirect wind Closure-composite rules • Super-secondary structures are composed of secondary structures that are neighboring in the chain sequence • However from the presence of a super-secondary structure, like a hairpin, in a protein structure follows that residues that are non consecutive become neighboring in space. Closure - “short cut” in the sequence due to a folding rule Example 1 applying folding rules to jelly roll Recursive domains Recursive domain is a part of a protein fold that can be generated using folding rules supported with the closure operation. A protein that can be fully generated using folding rules has one recursive domain. Examples • Example 1 • Example 2 • Example 3 • Example 4 Recursive domains Recursive domain is a part of a protein fold that can be generated using folding rules supported with the closure operation. A protein that can be fully generated using folding rules has one recursive domain. Graph theoretical tools and recursive domains Fold graph: Vertices – strands Edges – two types: Neighbor edges: directed edges between strands that are neighbors in chain or vie the closure operation. Domain edge: edges between stands used in the same folding rule Recursive domains = connected component of the fold graph without neighbor edges. Can the rules generate all known folds? Comparison with the partition for computer generated set of all possible 8-strand sandwiches Partition into recursive components for small (<=10 strands) proteins Control Protein data Distribution of recursive domains in all sandwich like '"folds" recursive domains for proteins with at most 10 strands 3000 45 40 2500 number of folds 30 25 recursive domains 20 15 number of generated "folds" 35 2000 1500 Series1 1000 10 500 5 0 1 2 3 4 5 6 7 8 number of recursive domains 0 1 One recursive fold 2 3 4 5 number of recursive domains 6 7 8 Offenders Hedhehog intein domain Given a fold, is there a unique sequence of folding steps leading to it? Usually no. Usually there alternative sequences of folding steps leading to a construction of the same domain. Do such alternative folding sequences correspond to alternative folding pathways? Are the rules complete? Probably not. e.g.: For propeller, each blade is in one recursive domain but we do not have a rule that will put the blades together. Conclusions: We are getting some idea how things work... It is so nice outside. It would be nice to take the dog for a walk! Nice… dog… walk Conclusions • Protein folds can be described by simple folding rules. • The folding rules capture at least some aspects of fold simplicity and regularity. • The sequence of folding steps leading to a given fold is usually not unique. • The folding rules generate protein-like structures. Future directions • Can folding rules guide fold prediction? • Would hierarchical description of a fold provided by folding rules be useful for fold classification / comparison ? • Adding statistical evaluation of a recursive domain. Acknowledgments George Rose Raj Srinivasen Rohit Pappu Venk Murthy NIH, K01 grant