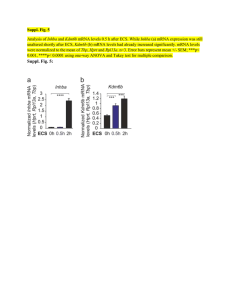
The Simpson-Elsasser-Wolfram (SEW) Framework for Modeling the Living Cell
advertisement

The Simpson-Elsasser-Wolfram Framework for Modeling the Living Cell Sungchul Ji Department of Pharmacology and Toxicology, Rutgers University sji@rci.rutgers.edu (Seminar at The DIMACS Center, Rutgers University, 4/17/2006) The Simpson-Elsasser-Wolfram Framework for Modeling the Living Cell • Introductory Remarks • Simpson, Elsasser, & Wolfram All Laws, Principles, and Concepts Apply. Physics and Chemistry are Necessary but Not Sufficient. Complex Systems Can Be Modeled with Simple Programs on a Computger. • The Bhopalator Dissipative Structures Conformons • Application to Budding Yeast Transcript Levels (TL) Tranascription Rate (TR) Transcript Degradation Rate (TD) • Conclusion Structure Conformons Energy Dissipative Structures (TL = aTR - bTD ) Function Budding Yeast, Saccharomyces cerevisiae Developmental options of S. cerevisiae (budding yeast) cell type. [H. Madhani, PNAS 97(25):13461-13463 (2000)]. The Living Cell The Cytoskeleton (Mouse Embryonic 3T3 cell) The Simpson-Elsasser-Wolfram Framework George G. Simpson Walter Elsasser (1902-1984) ( 1904 - 1991) Steven Wolfram (1959 - ) • G. Simpson: Physicists study the principles that apply to all phenomena: Biologists study phenomena to which all principles apply. • S. Wolfram: All structures and phenomena, whether internal or external, can be modeled on the computer as fractals, namely, the structures and patterns that emerge as the consequences of iterating the application of sets of simple rules n times, where n can be a large number (103 - 106). • G. Simpson: Physicists study the principles that apply to all phenomena: Biologists study phenomena to which all principles apply. Additional Principles: 1. General relativity & metabolic spacetime (H. A. Smith & G. R. Welch, 1991) “Just as matter tells spacetime how to curve and curved spacetime tells matter how to move, so biopolymers tells chemistry how , when and where to proceed and spatiotemporally organized chemical concentrations (IDSs) tell biopolymers how to move.” 2. Double articulation (1997) Covalent & noncovalent interactions 3. Rule-governed creativity (1997) Novel behaviors of cells are possible although cells depend on a finite number of chemicals and rules of interactions. 4. Semiotics of Peirce (2005) Biopolymers are molecular signs used in cell language 5. Soft-state physics (2006) Biopolymers are soft-state semiconductors of conformons. [S. Ji, BioSystems 44:17-39 (1997) ] 6. Biopolymers are coincidence detector s implementing the rule: If A and B, then C. (2006) DNA as the carrier of both genetic information and free energy. Figure 1. Supercoiled DNA duplexes. (Above) The electron micrograph of two circular DNA duplexes, one supercoiled into a compact shape and the other relaxed. (Right) Three shapes of DNA duplexes – a linear form (left), a circular form with one strand nicked (or cut) (middle), and a circular form that is closed and supercoiled (right). Visualization of Conformons Conformons = Mechanical energy stored in biopolymers as conformational strains localized at sequence-specific sites. The relaxed and supercoiled conformations of DNA: Supercoiled DNA harbors mechanical strains at sequence-specific sites, i.e., conformons . L. Stryer, Biochemistry, Fourth Ed., W. H. Freeman and Co., New York, 1995, p. 795 The Flame of a Candle as a Dissipative Structure • Like the Belousov-Zhabotinsky reaction run in a petri dish, the flame of a candle is a self-organizing chemical reaction-diffusion system, or a dissipative structure. Different reactions characterized by differently colored chemical species and temperatures occur in different regions in space. • The only major difference between the self-organizing chemical reactiondiffusion processes occurring in a candle flame and those proceeding in a living cell is that the former occur at high temperatures and the latter at low temperatures due to enzymic catalysis. • Therefore, the living cell can be viewed as supporting a cold flame vis-à-vis a hot flame of a candle. Thermal energy gradients and accompanying chemical concentration gradients in a candle flame • A candle flame embodies regions with different temperatures, ranging from 600 to 1,200 ºC. • Different temperatures are due to different chemical reactions proceeding in different regions in space that involve intermediates exhibiting different colors. Self-Organization • • • • The proto-typical example of selforganization is provided by the BelosovZhabotinsky (BZ) reaction discovered by the Russian chemist, B. P. Belousov in 1958. A few years later A. M. Zhabotinski confirmed and further elaborated on the the reaction. The BZ reaction is characterized by the organization of chemical concentrations in space (2- and 3-dimensions) and time (i.e., oscillating concentrations). The spatial patterns of chemical concentrations can evolve with time. ‘Patterns of chemical concentrations’ is synonymous with ‘chemical concentration gradients’. The organization of chemical concentration gradients in space and time in the BZ reaction is driven by the exergonic (i.e., free energy-releasing) chemical reactions constituting the BZ reaction itself. The Living Cell as a Dissipative Structure • The intracellular calcium ion gradient (an example of intracellular dissipative structures, IDSs) human neutropohils during phagocytosis [D. W. Sawyer et al., Science 230:663-666 (1985)] The florescent indicator called Quin2 is used to visualize intracellular calcium ion in human neutrophils during chemotaxis towards, and phagocytosis, of opsonized (i.e., coated with proteins that promote phagocytosis, or ingestion by cells) zymosan particles. • The pictures in the first column are brightfield images. The pictures in the second column are fluorescent images showing intracellular calcium ion distribution (white = high calcium; dark = low calcium). The pictures in the third column represent the color-coded ratio images of the same cells as in the second column. • Cells on the first row = Unstimulated neutrrophils. Cells on the second row = Neutrophils migrating toward an opsonized particle. Cells on the third row = Neutrophils with pseudopods surrounding an opsonized particle. Cells on the fourth row = Neutrophils after having ingested several opsonized particles. Take-home lesson = Dissipative structures in the form of ion gradients can form inside the cell without any membranes. The Action Potential as a Dissipative Structure • • • • The action potential is the transient depolarization of the neuronal membrane due to the equilibration followed by active transport of ions (Na+, K+, Ca++) across cell membrane in appropriate directions. The action potential is a heritable property of the neuron and hence must be encoded in the nucleotide sequences of DNA. How is such a dynamic properties of the cell encoded in the nucleus and how such encoded information is retrieved when needed are fundamental questions not yet fully answered. Three different kinds of ion movements are involved in the generation of the action potential: (1) primary active transport, (2) secondary active transport, and (3) passive transport. The Mikula-Niebur equation for the output rate of a coincidence detector [S. Mikula and E. Niebur, “The Effects of Input Rate and Synchrony on a Coincidence Detector: Analytical Solution. Neural Computat. 5(3):539-547 (2003)] “The coincidence detector is a computational unit that fires if the number of input spikes received within a given time bin equals or exceeds the threshold, Θ .” Nout = the output rate, firing per mint. p = the probability of a spike occurring within a time bin Δt. m = the number of input spike trains, each having n time bins. Θ = the threshold number of spikes that must be exceeded by the summed input spikes before the coincidence detector fires or is activated. q = the correlation coefficient between spike trains 1, . . . , m. j = the number of coincident spike trains. The Enzyme as a Coincidence Detector S. Ji, Ann. N. Y. Acad. Sci. 227:419-437 (1974) The Ca++ Ion Pump as a Coincidence Detector 20 1 / \ / \ 2 / \ / 4 / \ / 21 3 / \ \ / 5 6 \ 7 22 9 10 11 12 13 14 15 23 \ 8 / \ / 16 \ 17 18 19 20 21 22 23 24 25 . . . 24 P = mCj pj (1 – p)m-j , where mCj = m!/j!(m – j)! Figure 1. A temporal hierarchy composed of a set of coincidence-detecting events (CDEs), each constructed from a set of three nodes, j-i-k, where the simultaneous occurrence of events j and k leads to (or causes) event i. • W. Elsasser: Physicists study objects that belong to pure classes to which mathematical methods can be applied; biologists study objects that belong to heterogeneous classes to which logic, but not mathematics, can be applied. A Comparison between the Living Cell and the Atom 0.1 nm Dissipative Structure Equilibrium Structure Table 1. The different levels of description (granularity) involved in modeling the living cell Granularity Objects Number Emergent Properties Discipline Methods VII Cells 1030 ? Biological Functionality Cell Biology & Computer Science Wolfram’s Simple programs (Algorithmic Math?) VI Networks of Complexes 1020 ? Dissipative Structures & Mol. Logic Holistic Biology Bioinformatics (Discrete Mathematics) V Complexes 1015 ? Enhanced Coincidence Detection Biochemistry & Molecular Biology X-ray, TEM, SEM, etc. IV Macromolecules 1012 ? Coincidence Detection & Information Biochemistry & Molecular Biology Single Molecule Mechanics (C.M.) III Small Molecules 109 ? Diversity Chemistry Chemical Kinetics (C.M.) II Atoms ~ 102 Diversity Quantum Mechanics Molecular Dynamics (C.M.) I Subatomic Particles (e-, p+, n) 3 - Quantum Mechanics Molecular Dynamics (Cont. Math.) • S. Wolfram: All structures and phenomena, whether internal or external, can be modeled on the computer as fractals, namely, the structures and patterns that emerge as the consequences of iterating the application of sets of simple rules n times, where n can be a large number (103 - 106). Shell shapes generated by the simple model and found in nature. Two parameters are systematically changed: (a) the overall factor by which the size increases in the course of each revolution; (b) the relative amount by which the opening is displaced downward at each revolution. [S. Wolfram, A New Kind of Science, 2002, p. 416.] The Bhopalator Self-Organizing Chemical Reaction-Diffusion System = City Name + ator . (e.g., the Brusselator = the Belousov-Zhabotinsky reaction) The first molecular model of the living cell. Proposed in an international biological conference held in Bhopal, India, in 1983. The cell as a coincidence detector of coincidence detectors (2006) [S. Ji, J. theoret. Biol. 116:399-426 (1985)] Output Input 10 9 Gradients 5 6 4 Proteins Amino Acids 3 7 2 RNA Ribonucleotides 1 8 Genes A Simplified Representation of the Bhopalator [S. Ji, J. theoret. Biol. 116:399-426 (1985)]. DNA Microarrays • There are two kinds of DNA microarrays – cDNA or EST microarray and the Gene Chips. cDNA/EST microarrays are discussed first. • One microarray can measure 104 mRNA levels simultaneously • Each square can recognize one kind of mRNA molecules. • mRNA levels in the cell are determined by mRNA synthesis (Vsyn) and mRNA hydrolysis (Vhyd), because the rate of change in mRNA levels inside the cell is always: dR/dt = Vsyn - Vhyd • Only when certain kinetic conditions are met (to be discussed later) can the DNA microarray technique measure rates of gene expression. How DNA Microarray Experiments are Done 1 1. 2. 3 2 3. 4 4. 5. 5 6. 6 5 Isolate mRNA from broken cells. Synthesize fluorescently labeled cDNA from mRNA using reverse transcriptase and fluorescent nucleotides. Prepare a microarray either with EST or oligonucleotides (synthesized right on the microarray surface by Affimetric,Inc.). Pour the fluorescently labeled cDNA preparations over the microarray surface to effect hybridization. Wash off excess debris. Measure fluorescently labeled cDNA using a computer-assisted microscope. The final result is a table of numbers, each number registering the fluorescent intensity which is in turn proportional to the concentration of cDNA (and ultimately mRNA) located at row x and column y, row indicating the identity of genes, and y the conditions under which the mRNA levels are measured. Cluster Analysis The changes in mRNA levels of human fibroblasts (cells of connective tissues that synthesize and secrete fibrillar procollagen, fibronectin, and collagenase) measured with DNA microarrays over a time period of 24 hours. Green represents a decrease in mRNA levels (or “genes”, which term is strictly speaking incorrect; see below), black no change, and red an increase. Each mRNA molecule is represented by a single row of colored boxes, and each column represents measuring time point. Notice that the mRNA molecules belonging to cluster A started to decrease around 12 hours after beginning experiment. The mRNA molecules belonging to cluster E began to increase at around 8 hours after the beginning of the experiment. It is postulated that mRNA clusters represent dissipative structures of Prigogine produced by time-correlated activities of transcriptosomes and degradasome. A 1.2 1 1 0.8 0.6 0.4 0.2 0 6 0 fTL-fTR Plot 3 1.2 1 1 1 0.8 0.6 0.4 6 0.2 0 0.5 1 1.5 Fold Changes in TL 0 2 4 Fold Changes in TL D fTL-fTR Plot 10 C 6 fTL-fTR 19 2 1.2 1 Fold Changes in TR Fold Changes in TR B Fold Changes in TR Fold Changes in TR fTL-fTR Plot 1 1 0.8 0.6 6 0.4 0.2 6 1.5 1 1 0.5 0 0 0 1 2 Fold changes in TL 3 0 0.5 1 1.5 Fold Changes in TL Plots of fold changes in transcription rates (TR) and transcript levels (TL) of budding yeast during metabolic transitions caused by glucose-galactose shift. These three examples (for genes #1, #3, #10, and #19) were chosen randomly out of the 5,184 genes investigated by Perez-Ortin and his coworkers in Valencia, Spain. Each plot shows the results of 6 measurements at t = 0, 5, 120, 360, 450, and 850 minutes after glucose was replaced with galactose in the growth medium. Tim e - v_S plots = Glycolysis; = Oxphos Average m RNA Levels = Glycolysis; = Oxphos b mRNA, molecules/cell a 50 30 20 10 0 -200 1 0.8 0.6 0.4 0.2 0 v_S, molecules/cell/min 40 0 200 400 600 800 -200 1000 0 200 400 600 800 1000 Tim e, m in Tim e, m in Degradation/Transacription (D/T) Ratios vs Tim e = Glycolysis; = Oxphos c D/T Ratios 4.0 3.0 2.0 1.0 0.0 0 200 400 600 800 Tim e, m in Figure 1. The kinetics of mRNA level and transcription rate changes following glucose-galactose shift. (a) The time courses of the glycolytic and respiratory mRNA level changes. (b) The time courses of the glycolytic and respiratory transcription rate changes. (c) The time courses of the changes in the transcript degradation (D) to transcription (T) ratios. mRNA-Processor as a Coincidence Detector mRNA Synthesis (dnS/dt) Transcriptosome (> 50 proteins) 0<T<a Degradasome mRNA Level (dn/dt) (~ 10 proteins?) mRNA Degradation (dnD/dt) Dissipative Structures Cell Functions 1. Steady state (dn/dt = 0), if dnS/dt = dnD/dt 2. On-the-way-up state (dn/dt >0), if dnS/dnD > dnD/dt 3. On-the-way-down state (dn/dt <0), if dnS/dt < dnD/dt Table 9. An analogy between atomic physics and cell biology based on the similarity between line spectroscopy in atomic physics and cDNA array technology in cell biology. Parameter Atomic Physics Cell Biology 1. Time 19th-20th Century 20th-21st Century 2. Experimental Technique Atomic Absorption/Emission Spectroscopy (19th C) cDNA Array Technology (‘ribonoscopy’ ?)(1995) 3. Experimental Data Atomic Line Spectra mRNA Levels in the Cell 4. Regularities Lyman Series Balmer Series Ritz-Paschen Series Brackett Series Pfund Series RNA Metabolic Modules (‘ribons’) Genetic networks Cell Metabolic Networks 5. Theoretical Model Bohr’s Atom (1913) The Bhopalator (1985) 6. Basic Concepts Quantum of Action (1900) The Conformon (1972) IDSs (1985) Cell LanguageTheory (1977) 7. Theory Quantum Theory (1925) The Conformon Theory of Molecular Machines (1974) Cell Language Theory (1997) Molecular Information Theory (2004) 8. Philosophy Complementarity (1915) Complementarism (1993) 9. A Unified Theory of Physics, Biology, and Philosophy The Tarragonator (2005) [For references, see http://www.grlmc.com, under Publications]