Table S4
advertisement

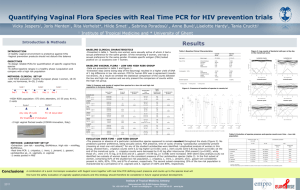
Table S4 The abundance of vaginal bacteria relative to total Bacteria gene copy number by species-specific qPCR for clinical cured participants in the 5-day follow-up after treatment a Group Species-Specific qPCR a BV-M (n=22) BV-L (n=25) p valueb Mean Std. Deviation Mean Std. Deviation Lactobacillus genus 76.98% 16.18% 58.74% 26.59% 0.1830 L. crispatusc 3.45% 10.90% 11.86% 24.31% 0.0000 L. inersc 60.75% 35.14% 44.89% 29.19% 0.0000 L. jenseniic 2.00% 2.86% 1.74% 3.56% 0.6157 G. vaginalis 0.00% 0.00% 2.96% 4.14% 0.0356 A. vaginae 0.00% 0.01% 0.00% 0.00% 0.1371 Eggerthella 0.00% 0.00% 0.00% 0.00% 0.2863 Megasphaera type 1 0.00% 0.00% 0.37% 1.72% 0.0300 Leptotrichia/Sneathia 0.00% 0.00% 0.00% 0.00% 0.1950 Prevotella 0.051% 0.13% 0.04% 0.16% 0.7325 The copy numbers of total bacteria were 2.77×108±2.58×108 copies/l in BV-M group and 7.40×109±8.18×109 copies/l in BV-L group respectively (p < 0.05). b Comparisons between women with BV and women without BV were calculated with independent-samples T-tests (SPSS Data Analysis Program version 16.0, SPSS Inc, Chicago, IL) and were considered statistically significant if p < 0.05. c The relative abundance of L. crispatus, L. iners and L. jensenii was compared to the copy number of Lactobacillus genus. 1