Table S1 Gene name
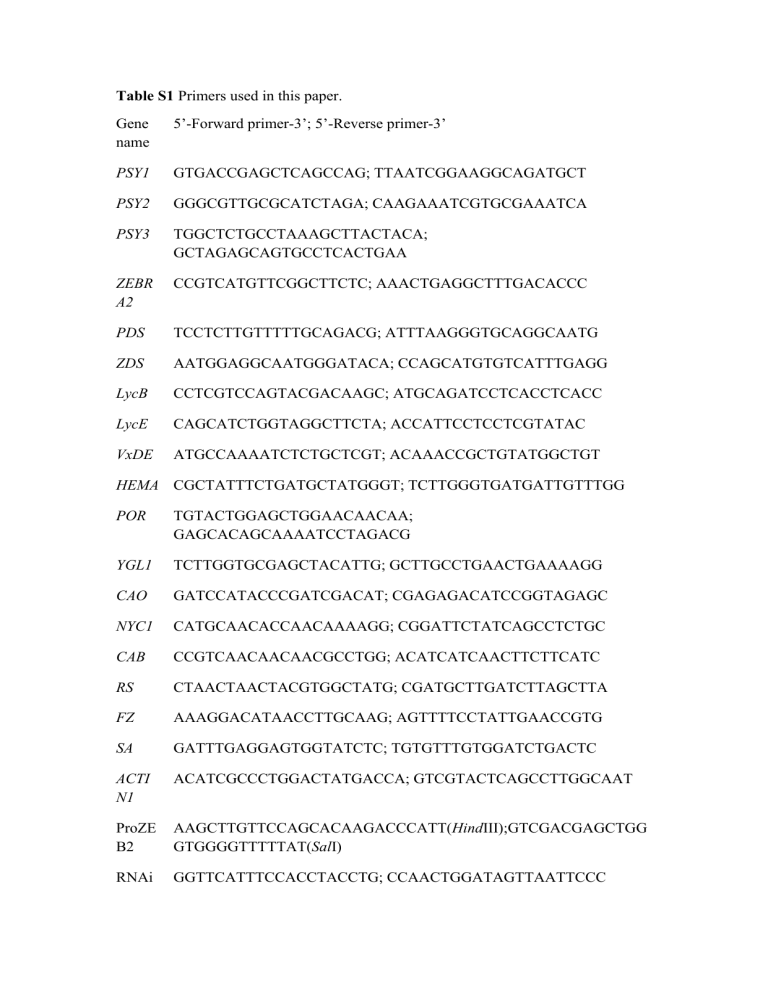
Table S1 Primers used in this paper.
Gene name
5’-Forward primer-3’; 5’-Reverse primer-3’
PSY1 GTGACCGAGCTCAGCCAG; TTAATCGGAAGGCAGATGCT
PSY2 GGGCGTTGCGCATCTAGA; CAAGAAATCGTGCGAAATCA
PSY3 TGGCTCTGCCTAAAGCTTACTACA;
GCTAGAGCAGTGCCTCACTGAA
ZEBR
A2
CCGTCATGTTCGGCTTCTC; AAACTGAGGCTTTGACACCC
PDS TCCTCTTGTTTTTGCAGACG; ATTTAAGGGTGCAGGCAATG
ZDS AATGGAGGCAATGGGATACA; CCAGCATGTGTCATTTGAGG
LycB CCTCGTCCAGTACGACAAGC; ATGCAGATCCTCACCTCACC
LycE CAGCATCTGGTAGGCTTCTA; ACCATTCCTCCTCGTATAC
VxDE ATGCCAAAATCTCTGCTCGT; ACAAACCGCTGTATGGCTGT
HEMA CGCTATTTCTGATGCTATGGGT; TCTTGGGTGATGATTGTTTGG
POR TGTACTGGAGCTGGAACAACAA;
GAGCACAGCAAAATCCTAGACG
RS
FZ
SA
ACTI
N1
YGL1 TCTTGGTGCGAGCTACATTG; GCTTGCCTGAACTGAAAAGG
CAO GATCCATACCCGATCGACAT; CGAGAGACATCCGGTAGAGC
NYC1 CATGCAACACCAACAAAAGG; CGGATTCTATCAGCCTCTGC
CAB CCGTCAACAACAACGCCTGG; ACATCATCAACTTCTTCATC
CTAACTAACTACGTGGCTATG; CGATGCTTGATCTTAGCTTA
AAAGGACATAACCTTGCAAG; AGTTTTCCTATTGAACCGTG
GATTTGAGGAGTGGTATCTC; TGTGTTTGTGGATCTGACTC
ACATCGCCCTGGACTATGACCA; GTCGTACTCAGCCTTGGCAAT
ProZE
B2
AAGCTTGTTCCAGCACAAGACCCATT( Hind III);GTCGACGAGCTGG
GTGGGGTTTTTAT( Sal I)
RNAi GGTTCATTTCCACCTACCTG; CCAACTGGATAGTTAATTCCC
Table S2 Relative abundance of individual carotenoid in the light-grown complemented (Comp.) line or zebra2-1 mutant compared with the wild type (WT) plant. The relative carotenoid abundance was shown as the ratio of peak area
(between the Comp. and WT or between zebra2-1 mutant and WT) which was derived from the HPLC chromatogram recorded at 430 nm of the light-grown WT, zebra2-1 mutant and Comp. lines.
Compound Peak area ratio neoxanthin
Comp./WT zebra2-1 /WT
0.85±0.13 1.07±0.21 violaxanthin
0.83±0.11 0.83±0.16
0.86±0.12 0.46±0.09 lutein
α-carotene
Zeaxanthin
0.78±0.08 0.30±0.05
β-carotene isomer 1 0.82±0.24 1.02±0.17
β-carotene isomer 2 0.83±0.15 1.33±0.13
0.77±0.14 3.82±0.11
Table S3 Relative abundance of individual carotenoid in the etiolated complemented
(Comp.) line or zebra2-1 mutant compared with the wild type (WT) line. The relative carotenoid abundance was shown as the ratio of peak area (between the Comp. and
WT or between the zebra2-1 mutant and WT) which was derived from the HPLC chromatogram recorded at 430 nm of etiolated WT, zebra2-1 mutant, and Comp. lines.
Compound Peak area ratio neoxanthin
Comp./WT zebra2-1 /WT
1.00±0.12
- violaxanthin
1.00±0.08
-
0.80±0.11
- lutein
ζ-carotene isomer 1 1.01±0.12 ∞
ζ-carotene isomer 2 1.00±0.13 44.67±3.12 prolycopene -
∞ cis -lycopene isomer 1
1.09±0.20 11.68±1.08 neurosporene isomer 1 0.98±0.13
∞ cis -lycopene isomer 2
0.93±0.18 18847.48±1465.43 neurosporene isomer 2 1.05±0.19
11844.81±1556.98
Table S4 Relative abundance of individual carotenoid in mature stems from the complemented (Comp.) line or zebra2-1 mutant compared with the wild type (WT) line. The relative carotenoid abundance was shown as the ratio of peak area (between the Comp. and WT or between the zebra2-1 mutant and WT) which was derived from the HPLC chromatogram recorded at 430 nm of mature stems from WT, zebra2-1 mutant, and Comp. lines.
Compound Peak area ratio zebra2-1 /WT
ζ-carotene isomer 1 8.87±0.96
ζ-carotene isomer 2 1081.17±141.11 prolycopene
∞ cis -lycopene isomer 1
27±0.07 neurosporene isomer 1 37.34±4.82 cis -lycopene isomer 2
∞ neurosporene isomer 2 108.11±9.50
Table S5 Relative abundance of individual carotenoid in leaves from the zebra2-1 mutant under field light condition or shaded compared with the wild type (WT) line.
The relative carotenoid abundance was shown as the ratio of peak area (between the zebra2-1 mutant and WT or the shaded zebra2-1 mutant and WT) which was derived from the HPLC chromatogram recorded at 460 nm of leaves from the WT and the zebra2-1 mutant (either under field light condition or shaded) plants.
Compound Peak area ratio neoxanthin zebra2-1 /WT shaded zebra2-1 /WT
0.70±0.16 1.01±0.35 violaxanthin
0.53±0.08
0.23±0.09 lutein
β-carotene isomer 1 0.94±0.09
0.83±0.13
0.38±0.12
0.99±0.22
β-carotene isomer 2 0.51±0.08 zeaxanthin
0.89±0.14
12.21±0.14 8.65±0.09
Fig. S1 Phenotypes of the wild type (WT), zebra2-1 mutant, complementary transgenic (Comp.) and RNAi transgenic (RNAi) seedlings at tillering stage.
Fig. S2 The CRTISO transcripts of different sizes detected in the wild type, zebra2-1 mutant and complemented plants. The specific 326-bp transcript in the wild type (lane
1), 302-bp transcript from zebra2-1 mutant (lane 2), and both 326-bp and 302-bp transcripts in the complemented line (lane 3) were detected by semi-quantitative RT-
PCR.
Fig. S3 CRTISO expression in the wild type, zebra2 mutant, complemented (Comp.) and RNAi (RNAi) lines. Quantitative RT-PCR was performed to quantify relative gene expression levels from at least biological triplicate for those lines.
Fig. S4 Absorbance spectra of several carotenoids in this study. (1) lycopene; (2) neoxanthin; (3) violaxanthin; (4) β-carotene; (5) α-carotene; (6) zeaxanthin; (7) lutein;
(8) prolycopene.