Trichosporon montevideense
advertisement

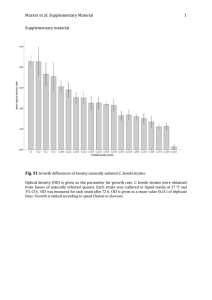
Supplementary Information for Biotechnology Letters Green synthesis of gold nanoparticles by a newly isolated strain Trichosporon montevideense for catalytic hydrogenation of nitroaromatics Wenli Shen, Yuanyuan Qu*, Xiaofang Pei, Xuwang Zhang, Qiao Ma, Zhaojing Zhang, Shuzhen Li and Jiti Zhou State Key Laboratory of Fine Chemicals, Key Laboratory of Industrial Ecology and Environmental Engineering (Ministry of Education), School of Environmental Science and Technology, Dalian University of Technology, Dalian 116024, China * Corresponding author. E-mail address: qyy@dlut.edu.cn (Y.Y. Qu) Materials and methods Chemicals and medium Nitroaromatics and aromatic ammonia compounds were obtained from Sinopharm Chemical Regent Beijing Co., Ltd. (China), including 2 -NP, 3-NP, 4-NP, o-NPA, m-NPA, 2-aminophenol, 3-aminophenol, 4-aminophenol, o-diphenylamine and m-diphenylamine. Hydrogen tetrachloroaurate (III) hydrate (HAuCl 4 ·3H 2 O) was purchased from J&K Scientific Ltd. (China). All other chemicals were of analytical grade or above. The medium used in selection and cultivation was lysogeny broth medium (LB, pH 7.0), which consisted of 10 g NaCl/l, 5 g yeast extract/l and 10 g peptone/l. Solid medium was obtained by supplement with 2% (w/v) agar. All the media were autoclaved at 121 oC for 20 min. Tetracycline (50 mg/l) and terramycin (50 mg/l) were added to the medium before use in the process of isolation strains. Identification of strain WIN Genomic DNA of strain WIN was applied to amplify the 26S rRNA gene by polymerase chain reaction (PCR), and PCR products were sequenced by TaKaRa Biotechnology Co. Ltd. (Dalian, China). The BLAST program was taken to compare the sequence with those in GenBank, and the related sequences were aligned using Clustal X (1.8). The aligned sequences were used to construct phylogenetic tree using Neighbor-joining method by MEGA (Version 5.1) with 1000 bootstrap replicate. Results and discussion Isolation and identification of strain WIN Strain WIN was spheroidicity as shown in SEM image ( Supplementary Figure 1a), and generated round colonies with centric points on agar plates (Supplementary Figure 1b). Based on the analysis of 26S rRNA gene sequence, strain WIN exhibited 99% homology to Trichosporon montevideense (Genebank accession number JQ965893), thus it was identified as Trichosporon montevideense. The phylogenic tree demonstrating the relationship between strain WIN and other strains was shown in Supplementary Fig. 1c. The partial 26S rRNA gene sequence (550 bp) of strain WIN was deposited in GenBank database under the accession number KP676895. Str ain WIN has been deposited in the China General Microbiological Culture Collection Center (CGMCC) under accession number CGMCC 10368. Supplementary Figure Supplementary Figure. 1 The SEM image (a) and agar plate photo (b) of strain WIN. (c) Phylogenetic tree of Trichosporon montevideense WIN and related species