Flow Cytometry Core A Resource of the Section of Immunobiology, Department of

Flow Cytometry Core
A Resource of the Section of
Immunobiology, Department of
Internal Medicine, and the YCCC
Presentation Overview
• Flow cytometry background and technical review
• Yale Flow Core: history, capabilities, people, and operation.
• Use of flow cytometry to study memory
B cells in my own lab.
What is Flow Cytometry?
• The analysis of single cells, particles, or other discrete elements as they flow past one or more focused light sources based on reflected, scattered, or fluorescent light generated by those light sources.
Fluorescence signals
Focused laser beam
Flow Cell
Injector
Tip
Sheath fluid
Laser
Forward Angle Light Scatter
FALS Sensor
Laser
90 Degree Light Scatter
FALS Sensor
90LS Sensor
Laser
Fluorescence Detectors
FALS Sensor
Fluorescence detector
(PMT3, PMT4 etc.)
Fluorescence
Fluorescence Activated
Cell Sorting
488 nm laser FALS Sensor
Fluorescence detector
Computer analysis of detector data“gating” leads to + or - charging of droplet
+
Charged Plates-
Attract and focus droplets of opposite charge
-
Single droplets sorted into test tubes
Optical Filters
Dichroic Filter/Mirror at 45 deg
Light Source Transmitted
Light
Reflected light
Standard Band Pass Filters
630 nm BandPass Filter
White Light Source
Transmitted Light
620 -640 nm Light
Standard Long Pass Filters
Light Source
520 nm Long Pass Filter
Transmitted Light
>520 nm
Light
Standard Short Pass Filters
Light Source
575 nm Short Pass Filter
Transmitted Light
<575 nm
Light
Flow cell
Flow Cytometry Optics
PMT
4
Dichroic
Filters
PMT
1
Bandpass
Filters
PMT
2
Laser
PMT
3
What can flow be used for?
• Expression of cell surface or intracellular proteins or neo-epitopes such as phospho-proteins generated via cell signaling (after permeabilization) using fluorescently tagged Abs.
• Use of small molecule, protein, or particle-based probes to detect:
Ca++ flux, pH, mitochondrial polarization (apoptosis surrogate), caspase activation, cell division via dye dilution (e.g. CFSE), phagocytosis, DNA content (cell cycle analysis) or proliferation
(BrdU incorporation).
• Detection of intrinsic cell fluorescence based on expression of fluorescent proteins from reporter constructs.
• Analysis of cell size and complexity using light scatter.
• Multiparameter analysis to determine cellular heterogeneity and to link properties to cell phenotypes in complex mixtures.
• Quantitative technique to enumerate specific cell types.
• Preparative method that can combine any of the above techniques isolate cells or even subcellular fractions at a rate of up to 50K events/second.
The Yale Flow Facility
History
• Began as 1 FACScan and FACStar plus, supported by HHMI and Section of Immunobiology.
• FACScalibur added in 94.
• FACSVantage in 98.
• FACS-DIVA upgrade in 2001-provided gratis by BD.
• Winter 2002: Current core opened in TAC with space and instruments contributed by Internal Medicine and purchase of additional used MoFlo by YSM. Custombuilt rooms.
• Initial configuration: 1 FACScan, 4 FACSCaliburs (3 from Int. Med), 2 sorters.
• Shared Instrument Grant for FACSAria in 2003.
• Fall of 2004: Purchase of LSRII-with UV capability for
DNA analysis.
• November 2004: YCCC merger completed.
Instrumentation
• FACS Analysis:
– 3 Color analysis: FACScan
– 4 Color analysis: 4 FACScaliburs
– 11-Color analysis: LSRII (new)
• Sorters:
FACSVantage DIVA: hi-speed digital stream in air sorter with UV, 488, 633; 4-way sort; cloning; aerosol containment.
MoFlo: hi-speed digital stream in air sorter with 488,
633; cloning
FACSAria: hi-speed digital cuvette sorter with 407,
488, 633; hi sensitivity, 13-color; 4-way sort; cloning; aerosol containment
7’-10”
TAC Building Room S617
7’-10”
Desk
4’-7”W x 2’-6”D
2’ W x
2’D
MoFlo
3’ D x 6’ W
7’-10”
FACS Vantage
4’ D x 3’ W
3’ W x
3’ D
10’ 10’
Scale 0.3”-1’ - Bench Space
31’
4’-7”
Lasers and Colors
Instruments
BD FACScan
BD FACSCalibur
BD FACSVantage
BD FACSAria
BD LSR II
Cytomation MoFlo
Laser
Argon (L1)
Argon (L1)
Red Diode (L2)
Argon (L1)
HeNe (L2)
Argon (L1)
HeNe (L2)
Violet (L3)
Argon (L1)
HeNe (L2)
UV (L3)
Violet (L4)
Argon (L1)
HeNe (L2)
Excitation Laser Line (nm) Fluorescence Channel
488 FL1 Green
FL2 Yellow
FL3 Red
488 FL1 Green
FL2 Yellow
FL3 Red
635 FL4 Red
488 FL1 Green
FL2 Yellow
FL3 Red
FL6 UV
633 FL4 Red
FL5 InfraRed
488 Green
Yellow
Red
Far Red
Infra Red
633 Red
Infra Red
407 Infra Red
488 Green
Yellow
Red
Far Red
Infra Red
633 FL4 Red
Infra Red
355 Violet
Blue
407 Blue
488 Green
Yellow
Red
Far Red
Infra Red
633 FL4 Red
Infra Red
Fluorochromes
FITC
PE
PE-Texas Red
FITC
PE
PE-Texas Red
APC
FITC
PE
PE-Texas Red
Hoechst
APC
APC-Cy7
FITC
PE
PE-Texas Red
PerCP-Cy5.5
PE-Cy7
APC
APC-Cy7
Alexa Fluor 405
FITC
PE
PE-Texas Red
PerCP-Cy5.5
PE-Cy7
APC
APC-Cy7
Alexa Fluor 405
Alexa Fluor 405
FITC
PE
PE-Texas Red
PerCP-Cy5.5
PE-Cy7
APC
APC-Cy7
Alexa Fluor 488
PI
PE-Cy5
Alexa Fluor 488
PE-Cy5
Alexa Fluor 647
Alexa Fluor 488
PI
PE-Cy5
Alexa 350
Alexa Fluor 647
Alexa Fluor 488
PE-Cy5
Alexa Fluor 647
Pacific Blue
Alexa Fluor 488
PE-Cy5
Alexa Fluor 647
Pacific Blue
Pacific Blue
Alexa Fluor 488
PI
PE-Cy5
Alexa Fluor 647
EMA
PerCP
PerCP
EMA
PerCP
Indo-1
PerCP
PerCP
EMA
PerCP
PerCP-Cy5.5
PE-Cy7
PerCP-Cy5.5
PE-Cy7
PerCP-Cy5.5
PE-Cy7
PI
PI
EMA
EMA
Capabilities and Techniques
• User operated 3, 4, and 11-color analysis.
• Technician-assisted analysis on request for an additional fee.
• Multicolor hi-speed digital sorting:
– 4-way
– Cloning/single cell or multicell
– Sterile
– UV, 407, 488, and 633 laser lines
– Operator performs most sorts; user-operation an option for experienced FACS Aria users.
• Techniques: cell surface markers, cytokines, intracellular staining, live-dead discrimination, Ca++-signaling,
DNA/cell cycle analysis, FRET, subcellular fractions, detection of almost any fluorescent molecule.
• Assistance with data analysis: two workstations with appropriate software.
Personnel
• Mark Shlomchik has run facility for the last 7 yrs. Currently my 5% effort is supported by a
PPG on which I am one of the PIs.
• Two sorter operators paid by effectively 50% by HHMI: Tom Taylor and Gouzel Tokmoulina.
• One R+D tech who supervises FACS Aria and analyzers: Geoff Lyon.
• One new R+D tech who oversees the analyzers, billing and training: Don Foster.
User Support and Resources
• Consultation on sorts, analysis, etc. before, during or after the experiment, as needed, no charge.
• Regular training sessions for all instruments.
• Broadcast announcements via email about new policies, unexpected downtime, etc.
• Sponsorship of training seminars on data analysis, new reagents, techniques.
• Negotiated discounts with a variety of vendors.
User Support and Resources
• New web site: designed and maintained by
Gouzel Tokmoulina.
– Description of equipment
– Facility rules
– Sign-up
– Resources and information
• Web-based scheduling for all analyzers.
Usage Statistics
• Analysis: ~950 hr./month
• Sorting: ~325 hr./month
• 119 Active laboratories
• >486 Registered individual users
Budget
• Sources of income
– User fees: ~$440K
– YCCC: $39K
– HHMI: ~$112K
• Costs
– Salaries: ~$300K/yr. Salary+fringe for 4 people and administrative support.
– Maintenance contracts: $135K/yr. (some deferred due to prepaid contracts.
– Other: ~$25K/yr.
Breakdown of Key Fees
• User-operated analysis: $14/hr.
• User-operated LSRII or Aria (analysis only): $22/hr.
• Operator-performed sorting or analysis:
$68/hr.
• Training: $30/hr. for up to three people
(1 hr. sessions).
Future Plans
• Additional LSRII in one to two years. We have applied for a Shared Instrument Grant that is pending review.
• Further application development and customer support and training.
• Expansion with a satellite facility in the
Amistad Building to support Stem Cell,
Vascular Biology and Human Immunology programs: will need a new sorter and an
LSRII analyzer. All users could use either facility.
• Expanded education mission.
• Ideas for what you want or need?
Applications Examples-Study of B cell Memory
• Identifying memory cells and phenotyping them using BrdU labeling.
• Sorting memory cells for mRNA or subsequent functional analysis.
• Using FACS to confirm the microarray data.
Definition of a “Memory Cell”
• A cell that has previously responded to antigen and that persists in a resting state for a long period of time after initial exposure.
• What are the features that distinguish resting “memory” and naïve B cells?
Membrane IgM (mIgM) frequency of hapten NIP-specific B cells
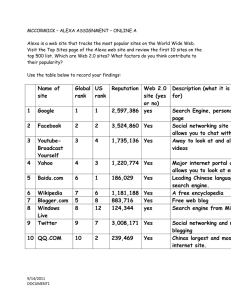
s
VDJ E switc h
C
M
Bl/6, V
H
186. 2 Balb/c IgM a, no secreted exo n
Signals for production of secreted IgM have been deleted .
Only membrane-bound IgM a is produced by the transgene.
Pairing of the V
H
186.2
variable region gene with endogenous
1 light chain produces antibody specific for the hapten NP.
This provides a system in which the functions of B cells and antibody can be distinguished.
Expansion of Antigen-specific
Population 12 weeks after
Immunization
Summary of NP+ Expansion after Immunization
BrdU Labeling Strategy
BrdU is administered intraperitoneally every 12 hours on the days indicated above.
BrdU-positive Ag-specific B cells present in mIg and (m+s)Ig mice
12 weeks post immunization
Decay Kinetics of BrdU-labeled
Memory B cells is Equivalent in mIg and (m+s)Ig mice
BrdU+NP+ Half life >8wks n=5-14 mice
BrdU-positive, NP+ B cells present in mIg and (m+s)Ig Immune mice were compared with NP+ cells in Alum control mice
Resting Memory Cells Have High
CD80 Expression
B o l d =immune
A.
A System for Generating Large Numbers of Memory Cells for Further Study:
Hyperimmunization of mIg mice
NP-CGG NP-CGG Assay
C.
- 4wk d0
B.
Naive
1.66
8 12 16 20wk
Two Doses
7.96
NP+ 12 weeks post 2nd immunization
APPROACH
Naïve or Immunized a
-NP Splenocytes
93.8
FACS Sorting
RNA Isolation
Labeled cRNA Preparation
Affymetrix Hybridization
Statistical Analysis
Data Mining
92.4
1.68
Naive
Memory
3.16
Kappa
Biological Replicates
#1
Naïve NP 3
Memory NP 4
GC NP
#2
4
4
6
Naive
FACS Isolation of NP+ Splenic B Cells for
Affymetrix and qPCR Analysis
4 o C; NaAzide 0.05%; FCS
50.8
97.3
35.1
Bonnie JhD-/- Jk-/- mice
PI
Memory
97.9
PI
Fitc-anti-B220
Fitc-anti-B220
Bonnie JhD-/- mice 12 weeks post 2nd immunization
51.7
APC-NIP
17.4
79.3
77.9
Biot-anti-Kappa
Cell Surface Molecules: M>N
(Affy and qPCR Confirmed
Mela (80-kD Melanoma Antigen)
Emp1 (epithelial membrane protein 1)
Bmpr1a (bone morphogenetic protein receptor, type 1A)
Atp11a (ATPase, class VI, type 11A)
Myadm (myeloid-associated differentiation marker)
Adora2a (adenosine A2a receptor)
CD80
CD36
Acknowledgements
• Tom, Geoff, Gouzel, and Don
• HHMI, Internal Medicine, YCCC
• YSM Administration
• Shannon Anderson and Mary Tomayko
• Today’s speakers
![Mouse IgG2b, kappa monoclonal [7E10G10] - Isotype](http://s2.studylib.net/store/data/012909847_1-9b2bb6a95a189600a77028a367bfe36d-300x300.png)

![Anti-CD147 antibody [EPR4053] (Alexa Fluor® 488) ab205450](http://s2.studylib.net/store/data/012963350_1-9f029359b62a58420c39721f185df4dd-300x300.png)
![Anti-BNIP3 antibody [ANa40] (Alexa Fluor® 647) ab196706](http://s2.studylib.net/store/data/012083394_1-2ff7db27c0d6912ecfc1f982c1a7d990-300x300.png)