Yi Lu Functional DNA Fluorescent and Colorimetric
advertisement

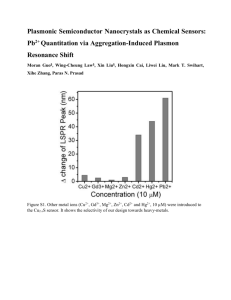
Functional DNA Fluorescent and Colorimetric Sensors for Heavy Metal ions and Endotoxins Yi Lu Department of Chemistry, Biochemistry, Bioengineering, Materials Science and Engineering, Center for Biophysics and Computational Biology, and Beckman Institute for Advanced Science and Technology University of Illinois at Urbana-Champaign Urbana, IL 61801 A Grand Challenge Facing Natural Resource Sustainability On-site, real-time detection and quantification of toxic materials, such as heavy metal ions and endotoxins, before, during and after remediation. A Major Scientific and Technological Issue Group 1 Period 2 1 1 H 2 Li Be 3 Na Mg 4 3 11 19 4 12 20 3 4 5 6 7 8 9 10 11 12 • Essential Metal ions • Bulk and essential non-metal ions • Non-essential biological elements • Toxic metal ions • Radionuclides 23 18 2 He 5 6 7 8 9 10 B C N O F Ne 13 14 15 16 17 18 Al Si P S Cl Ar 31 29 30 32 33 34 35 36 Zn Ga Ge As Se Br Kr 37 39 40 47 48 49 50 51 52 53 54 Ag Cd In Sn Sb Te I Xe 44 27 17 Cu 43 26 16 28 42 25 15 Sc Ti V Cr Mn Fe Co Ni 41 24 14 K Ca 38 21 22 13 45 46 5 Rb Sr Y Zr Nb Mo Tc Ru Rh Pd 6 Cs Ba * Lu Hf Ta W Re Os Ir 55 71 72 7 Fr Ra ** Lr Rf Db Sg Bh Hs Mt Uun Uuu Uub Uut Uuq Uup Uuh Uus Uuo 87 56 88 73 74 75 76 77 78 79 80 81 82 83 84 85 86 Pt Au Hg Tl Pb Bi Po At Rn 112 113 114 115 116 117 118 69 70 103 104 105 106 107 108 109 110 57 58 59 60 61 62 63 64 *Lanthanides * La Ce Pr Nd Pm Sm Eu Gd 89 90 91 92 93 94 95 96 111 65 66 Tb Dy Ho Er Tm Yb 98 99 101 Cf Es Fm Md No 97 **Actinides ** Ac Th Pa U Np Pu Am Cm Bk 67 68 100 102 Toxic materials in the environment are large in numbers, subtle in structural differences, small in quantities. Therefore they represent new challenges and opportunities in natural resource sustainability. Instrumental Analyses Mass spectrometry Inductively Coupled Plasma Advantages Industrial standard Highly sensitive (down to ~ppb or less) Detect a number of analytes simultaneously Disadvantages Require sophisticated equipment, samplepretreatment, and skilled operators difficult for in-situ, on-site, remote or real-time detection/imaging Four key steps in designing sensors ? a general method to obtain molecules for any specific target (e.g., Lead, mercury, cocaine, endotoxins, cancer) ? a general method to improve selectivity; a general method to transform molecular recognition into physical detectable signals without compromising the binding affinity and selectivity; ? ? a general method to fine-tune the dynamic range. Four key steps in designing sensors ? a general method to obtain molecules for any specific target (e.g., Pb2+, Amphetamine, cocaine, Ricin, cancer) virus Metal ions pH Method toxins bacteria Organic cofactors Until recently, antibody is the only method that is general enough for a broad range of targets, but antibodies are not very good at sensing small molecules. Functional DNA RNA DNA O 5' A O O DNA/RNA = Protein enzymes DNA/RNA = Antibodies A O 2' 3' H O O P O OH O O P O O C O Substrate C O M(II) A: DNAzyme O Enzyme H O OH O O P O + O P O G O G O O O H O O P O OH O O P O T O B: Aptamer U O O O H O O P O O C: Aptazyme OH O O P O O M(II) H H N N H N O N N N H N N N H O T N N N N A H N O N H O G C H Kruger, K. et al. Cell 31, 147 (1982). Guerrier-Takada, C. et al. Cell 35, 849 (1983). Breaker, R.; Joyce, G. Chem. Biol. 1, 223 (1994). Combinatorial biology: a general method to obtain DNA/RNA for a specific target In vitro Selection Systematic Evolution of Ligands by Exponential Enrichment (SELEX) Advantages: • High throughput (1014-1015 different sequences) • Selective Amplification • Improvement in each round • Minimal cost • Short time Ellington, A. D.; Szostak, J. W. Nature 346, 818(1990). Tuerk, C.; Gold, L. Science 249, 505 (1990). Beaudry, A. A.; Joyce, G. F. Science 257, 635 (1992). Molecules Recognized/bound by Selected DNA/RNA Analyte/target type Examples Metal ions Mg(II), Ca(II), Mn(II), Pb(II), Hg(II), U(VI) Organics Cibacron blue, reactive green 19 Amino acids L-Valine, D-Tryptophan Nucleosides/nucleotides Guanosine, ATP Nucleotide analogs 8-oxo-dG, 7-Me-guanosine Biological cofactors NAD, FMN, porphyrins, Vitamin B12 Aminoglycosides Tobramycin, Neomycin Antibiotics Streptomycin, Viomycin Peptides Rev peptide Enzymes Human Thrombin, HIV Rev Transcriptase Growth cofactors Karatinocyte GF, Basic fibroblast GF Antibodies human IgE Gene regulatory factors elongation factor Tu Cell adhesion molecules human CD4, selectin Intact viral particles Rous sarcoma virus, Anthrax spores Juewen Liu and Yi Lu, J. Fluoresc. 14, 343-354 (2004). Examples of in vitro selected DNAzymes 3' N N N N N N Y rR N N N N N N N 5' 5' N N N N N N R N N N N N N N 3' A G G G C C T A A A G C CTA 3' N N N N N N G rN N N N N N N N 5' 5' N N N N N N T N N N N N N N 3' AA C C G G G A G TC G C C Mg2+ Pb2+ UO22+ Zn2+ 5' Y Y Y Y A A T A C G N N N N N 3' R R R R A C 3' YYYY C CGG C N N N N N 5' T G Cu2+ Lu, Y. Chem. Euro. J. 8, 4588-4596 (2002). Co2+ Examples of in vitro selected DNAzymes 3' N N N N N N Y rR N N N N N N N 5' 5' N N N N N N R N N N N N N N 3' A G G G C C T A A A G C CTA 3' N N N N N N G rN N N N N N N N 5' 5' N N N N N N T N N N N N N N 3' AA C C G G G A G TC G C C UO22+ Mg2+ Pb2+ Substrate M(II) Enzyme 5' Y Y Y Y A A T A C G N N N N N 3' R R R R A C 3' YYYY C CGG C N N N N N 5' T G Cu2+ General secondary structure, making it easy for sensor design Lu, Y. Chem. Euro. J. 8, 4588-4596 (2002). Why using functional DNA in sensing? environmentally benign cost effective stable under rather harsh conditions DNA is ~1,000-fold more stable to hydrolysis than proteins and ~ 100,000fold more stable than RNA Globular shape: • More resistant to nucleases • Less likely to bind other molecules can be denatured and renatured many times (for manufacturing and storage) Non-immunogenic Can be readily modified for signal transduction can be genetically engineered to be delivered to a specific location Antibody Functional DNA Juewen Liu, Zehui Cao, and Yi Lu, Chem. Rev. 109, 1948-1998 (2009). Four key steps in designing sensors √ a general method to obtain molecules for a specific analyte; ? a general method to improve selectivity; What most want to do What most end up doing Problems with Selectivity in in vitro Selection 0.12 0.1 kobs (min-1) 0.5 mM Co(II) 0.08 0.5 mM Zn(II) 0.5 mM Pb(II) 0.06 0.04 0.02 0 No Negative Selection • • Sequences selected with Co2+ show high activity with Zn(II) and Pb(II) Must reduce or remove unwanted activity to improve metal selectivity Initial pool of DNA/RNA 2. A general method to improve selectivity: negative selection Selections using only Co 2+ DNA/RNA with 2+ Co activity Positive Selection Incorporate negative selection to reduce “unwanted” activity Can be performed in parallel with positive selection for comparison Selection using only Co 2+ Negative Selection Selection with a "metal soup" containing competing metal ions Remove sequences that are active with other metal ions 2+ High Co activity, 2+ Continue Co selection 2+ but with low Co P. J. Bruesehoff, J. Li, A. J. Augustine III, and Y. Lu, Combinatorial Chemistry and High Throughput Screening, 5, 327-335 (2002). selectivity 2+ High Co activity with 2+ high Co selectivity Improved Metal Ion Selectivity after Negative Selection 0.25 0.2 0.5 mM Co(II) kobs (min-1) 0.5 mM Zn(II) 0.15 0.5 mM Pb(II) 0.1 0.05 0 No Negative Selection Negative Selection P. J. Bruesehoff, J. Li, A. J. Augustine III, and Y. Lu, Combinatorial Chemistry and High Throughput Screening, 5, 327-335 (2002). Four key steps in designing sensors √ a general method to obtain molecules for a specific analyte; √ a general method to improve selectivity; ? a general method to transform molecular recognition into physical detectable signals without compromising the binding affinity and selectivity; Analytes Signal Target Signal recognition transduction part part DNAzyme/ Aptamers Fluorescence Color Turn analyte-dependent catalytic reactions into physically detectable signals A general method to convert DNAzymes into fluorescent sensors using catalytic beacon cleavage site substrate strand 17DS 3'- G T A G A G A A G G rN T A T C A C T C A -5' 5'- C A T C T C T T C T A T A G T G A G T -3' Rh-17DS A A C C G G T CG A G enzyme strand 17E C G C 17E-Dy COO3' 5' 17DS 5' 17E O O 3' O P TAMRA O O O N N N H OH N Dabcyl Fluorescent tag Hybridization with catalytical DNA Fluorescence suppressed rA Catalytic DNA strand Quencher Pb2+ rA 80000 fluorescence intensity rA Catalytic DNA strand H N O O P O O- O- RNA linkage Substrate strand N+ O N I III +Enz 60000 II I +Pb2+ 40000 III 20000 II 0 570 600 630 660 Quencher wavelength (nm) Li, J.: Lu, Y. J. Am. Chem. Soc., 122, 10466-10467. (2000). 690 A general method to convert DNAzymes into fluorescent sensors using catalytic beacon for a broad range of metal ions Fluorescent sensors Based on catalytic beacon Pb2+: Detection limit: 1 nM EPA MCL: 75 nM UO22+: Detection limit: 45 pM EPA MCL: 126 nM ICP-MS: 420 pM Cu2+: Detection limit: 35 nM EPA MCL: 20 μM Hg2+: Detection limit: 2.4 nM EPA MCL: 10 nM Li, J.: Lu, Y. J. Am. Chem. Soc., 122, 10466-10467. (2000). J. Liu, et al. Proc. Natl. Acad. Sci 104, 2056 (2007). J. Liu and Y. Lu J. Am. Chem. Soc. 129, 9838 (2007). J. Liu and Y. Lu, Angew. Chem., Int. Ed., 46,7587 (2007). High specificity or selectivity Over 1 million fold selectivity over Th(IV), and hundreds of millions fold selectivity over other metal ions Juewen Liu, Andrea K. Brown, Xiangli Meng, Donald M. Cropek, Jonathan D. Istok, David B. Watson, and Yi Lu, Proc. Natl. Acd. Sci. USA 104, 2056 (2007). Design of a Simple Colorimetric Biosensor cleavage site substrate strand 17DS 3'- G T A G A G A A G G rAPb T A T C A C T C A -5' 5'- C A T C T C T T C T A T A G T G A G T -3' A A C C G G T CG A G enzyme strand17E C G C BLUE Mix Aggregate Pb RED Pb 17E = 5'GGAAGAGATG Au = Pb BLUE = S rA -3' = SubAu 5'-CACGAGTTGACA-3' Pb RED = DNAAu Liu, J.; Lu, Y. J. Am. Chem. Soc. 125, 6642-6643 (2003). pH Indicator like Operation 0 0.3 0.5 5 M Mg(II) Ca(II) pH 9.5 Mn(II) 8.0 1 Co(II) 7.0 2 3 4 Ni(II) Cu(II) Zn(II) 4.0 3.0 2.0 Liu, J.; Lu, Y. J. Am. Chem. Soc. 125, 6642-6643 (2003). Liu J.; Lu, Y. Chem. Mater., 16, 3231 (2004); Liu J.; Lu, Y. J. Am. Chem. Soc. 126, 12298 (2004). 5 M Pb(II) Cd(II) 1.0 Colorimetric sensors for Uranyl Detection limit: 50 nM EPA MCL: 130 nM 1 nM Lee, J.-H.; Wang, Z.; Lu, Y. J. Am. Chem. Soc. 130, 14217-14226 (2008). Wang, Z.; Lee, J.-H.; Lu, Y. Adv. Mater., 20, 3263–3267 (2008). Four key steps in designing sensors √ a general method to obtain molecules for a specific analyte; √ a general method to improve selectivity; a general method to transform molecular recognition into physical detectable signals without compromising the binding affinity and selectivity; ? a general method to fine-tune the dynamic range. The need for tunable dynamic range • 22 million old houses in the US alone have used lead paint • US federal “thresholds” are 1.0 mg/cm2 and 0.5% • Current lead detection kits are based on Na2S9H2O or sodium rhodizonate. • A study of available kits showed low rates of both false positive and false negative results when compared to laboratory analytical results using the federal thresholds (www.hud.gov) • Current kits cannot tune the dynamic range Threshold yes Signal yes no no Lead concentration IDEAL CURVE Threshold A Colorimetric Biosensor with Tunable Dynamic Range A. Active DNA cleavage site substrate strand 17DS 3'- G T A G A G A A G G rA T A T C A C T C A -5' Extinction Extinction 1.5 1.5 3'- G T A G A G A A G G rA T A T C A C T C A -5' 5'- C A T C T C T T C C A T A G T G A G T -3' A A C C G G T CG A G enzyme strand17E C G C 0.5 0.5 0.0 0.0 15 15 C C 12 12 EE522 /E /E700 522 700 cleavage site substrate strand 17DS B B 450 450 550 650 750 450 550 550 650 650 750 750 450 550 650 750 Wavelength Wavelength Wavelength (nm) (nm) Wavelength (nm) (nm) 5'- C A T C T C T T C T A T A G T G A G T -3' A A C C G G T CG A G enzyme strand17E C G C B. Inactive DNA 1.0 1.0 A A 9 9 B:A = 20:1 B:A = 0:1 6 6 3 3 0 0 0.01 0.01 0.1 0.1 1 1 [Pb(II)] [Pb(II)] M M Brown, A. K.; Li, J.; Pavot, C. M.-B.; Lu, Y. Biochemistry 42, 7152-7161 (2003). Liu, J.; Lu, Y. J. Am. Chem. Soc. 125, 6642-6643 (2003). 10 10 100 100 Pb2+ Detection in Paint 1.0 mg/cm2 (threshold) qualitative 0 0.05 0.34 1.22 0.81 4.71 1.52 mg Pb / cm2 paint 10 8 Latex paint Oil paint 7 Latex on latex Oil on oil Oil on Latex Latex on Oil 8 A538/A700 A538/A700 6 5 4 3 6 4 2 2 1 Portable colorimeter quantitative 0 0 0 1 2 mg/cm2 Pb 3 4 5 0 1 2 3 4 2 mg/cm Pb 5 6 7 The tunable sensor allowed our Pb sensor to change color right at the federal threshold of 1.0 mg/cm2, and to work with different types of paints. Beyond metal and catalytic DNA-based detection Metal ions (Mg(II), Ca(II), Pb(II), Zn(II)) Organic dyes (Cibacron blue, reactive green 19) Amino acids (L-Valine, D-Tryptophan) Nucleosides/nucleotides (Guanosine, ATP) Nucleotide analogs (8-oxo-dG, 7-Me-guanosine) Biological cofactors (NAD, FMN, porphyrins, Vitamin B12) Aminoglycosides (Tobramycin, Neomycin) Antibiotics (Streptomycin, Viomycin) Peptides (Rev peptide) Enzymes (Human Thrombin, HIV Rev Transcriptase) Growth cofactors (Karatinocyte GF, Basic fibroblast GF) Antibodies (human IgE) Gene regulatory factors (elongation factor Tu) Cell adhesion molecules (human CD4, selectin) Intact viral particles (Rous sarcoma virus, Anthrax spores) Juewen Liu and Yi Lu, J. Fluoresc. 14, 343-354 (2004). Endotoxins • Endotoxins - potentially toxic natural compounds found in the outer cell membrane of various gram-negative bacteria • Also known as LPS (LipoPolySaccharides) and consist of a polysaccharide chain and a lipid moiety • Humans exposed to endotoxins either through ingestion and inhalation of gram negative bacteria either in medical environments or in industrial settings • Ailments including skin rashes, malaise, fevers and respiratory distress attributed to endotoxin exposure • USP and FDA standard is the Limulus Amebocyte Lysate (LAL) gel-clot method, which suffers from interferences due to large number of contaminants present in metalworking fluids, common in industrial environments Limulus polyphemus: commonly known as Horseshoe-crab Structure of Endotoxins (or LPS) a) Inner Core Outer Core O-Specific Chain General architecture : Lipid A Lipid Polysaccharide CO2- b) Structural representation : Kdo CO2GluN Dgalactose heptose PO4DGluN Kdo CO2- Dglucose DDgalactose glucose heptose PO4- Polysaccharides KDO : 2-keto-3-deoxyoctulosonic acid Present in LPS expressed by most gram-negative bacteria heptose PO4- Kdo DGluN PO4Lipid Selection progress 2.5 Switching Activity 2 Endotoxin KDO 1.5 1 0.5 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Selection Round • Switching Activity = Ratio of radiation of eluted fractions over background • Round 1-15 : small increase over background ; R16 : double the background Jennifer Deluhery and Nandini Nagraj A general method to turn aptamers into colorimetric sensors Liu J.; Lu, Y. Angew. Chem., Int. Ed., 45, 90-94 (2006). Liu, J.; Lu, Y. Nature Protocol 1, 246-252 (2006). A dipstick test using functional DNA nanoparticles Cocaine Juewen Liu, Debapriya Mazumdar and Yi Lu, Angew. Chem., Int. Ed. 45, 7955 –7959 (2006). Summary To obtain sensors for a broad range of targets, general strategies have been developed to • • • • to obtain sensing molecules; to improve selectivity; to convert molecular recognition event into physically detectable signals; to tune the dynamic range. The combination of functional DNA and nanotechnology resulted in sensors that • are stable and cost-effective. • are highly sensitive and selective. • have tunable dynamic range • can be applied to detection and quantification of a broad range of targets (metal ions, organic molecules, proteins and cells….) • allow real-time, on-site (or remote) sensing. Acknowledgments DNA/RNA Lab at UIUC Collaborators Dr. Jennifer Deluhery, Dr. Kishore Rajagopalan (ISTC) Graduate Students: Margaret Shyr, Dr. Paul Braun (MatSE, UIUC) Ying He Andrea K. Brown Abhijit Mishra, Gerard C. L. Wong (MatSE, UIUC) Hannah Ihms Peter J. Bruesehoff Rong Tong and Jianjun Cheng (MatSE, UIUC) Tian Lan Hee-Kyung Kim Tae-Jin Yim, Dr. Jonathan S. Dordick (CheE, RPI) Darius Brown Jing Li Carla B. Swearingen, Dr. Paul W. Bohn (Chem, UIUC) Nandini Nagraj Jung Heon Lee Kashan A. Shaikh, Dr. Chang Liu (ECE, UIUC) Eric Null Juewen Liu In-Hyoung Chang, Dr. Donald M. Cropek (CERL, DoD) Zidong Wang Kevin E. Nelson Brian Wong Daryl P. Wernette Funding: Weichen Xu Mehmet V. Yigit ISTC, EPA, HUD, NIH, DOE, NSF, & DOD Hang Xing Debapriya Mazumdar Yu Xiang Seyed-Fakhreddin Torabi Postdoctoral fellows: Hui Wei Zehui Cao Juanzuo Zhou Geng Lu Xiangli Meng Daisuke Miyoshi Wenchao Zheng Selection Strategy biotin 5' avidin-coated bead 5' 5' P1 P1 P2 annealing 1 5' 5' 5' 5' 5' annealing 5' 2 5 R 1. NaOH 2. PAGE P2 5' 1. Incubation 2. Elution 5' 5' 5' 5' 4 3 5' PCR 5' R 5' • Selection with 1.25 µg/mL endotoxin in 50 mM HEPES-Na at pH 7.35 , containing 50 mM NaCl, 5 mM KCl and 5 mM MgCl2 • Parallel positive selection with KDO to expand selectivity and sensitivity of aptamers to most gram-negative bacterial strains In vitro selection In vitro selection with [Mn+] as metal ion cofactor N40 PCR B rA N40 + rA B B Metal cofactor B Cloning and sequencing PCR Introducing mutations rA rA Further selection by decreasing reaction time and [Mn+] NaOH B rA Cloning and sequencing R. R. Breaker, G. F. Joyce Chem. & Biol. 1, 223 (1995). J. Li, W. Zheng, A. H. Kwon, and Y. Lu Nucleic Acids Res. 28, 481 (2000). Characterization This method is adaptable to any metal ion and any oxidation state of the same metal ion with tunable activity, and affinity In vitro selection Liu, J. et al. Proc. Natl. Acad. Sci 104, 2056 (2007). Future Goals • Continuation and comparison of selection with both endotoxin as well as KDO • Incorporation of negative selection with metalworking fluids to increase selectivity • Variation of incubation time to further increase sensitivity Selection Progress : Gel-based assays LPSR10 M KDOR10 PAGE gels after every round of PCR amplification to separate bound sequences from unbound sequences