Characterization of -Amyloid Aggregates and Aggregation Inhibition molecules using various techniques
advertisement

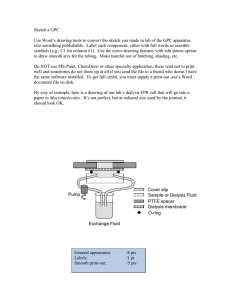
Macromolecules for The Demented and methods for their study Help from Keunok Yu, Jirun Sun, Bethany Lyles, George Newkome and LSU’s Alz-Hammer’s Research Team Krispy Kreme Donut Day, September 2003 Supported by National Institutes of Health-AG, NSF-DMR and NSF-IGERT • • • • How Alzheimer’s happens Attempts to prevent or reverse it Characterization challenges Alzheimer’s model systems with materials implications Amyloid Diseases • Several diseases are caused by the misfolding of proteins into self-associating structures (fibrils) w/ predominantly b-sheet secondary structure • Alzheimer’s Disease: Amyloid b-protein (Abin neurons/brain • Type II Diabetes: amylin in Islets of Langerhans • Mad Cow Disease (BSE): PrpSc in brain -- transmittable by protein aggregates • Huntington’s Disease, triplet repeat expansion: (Gln)n Age: Positron emission tomography 20 80 Normal 80 Alzheimer’s Postmortem Coronal Sections Normal Alzheimer’s PET images courtesy of the Alzheimer's Disease Education and Referral Center/National Institute on Aging; Postmortem images courtesy of Edward C. Klatt, Florida State University College of Medicine APP = Amyloid Precursor Protein http://www.bmb.leeds.ac.uk/staff/nmh/amy.html APP = the larger, lighter pink one •Transmembrane protein •Normal function not known •Educated guesses May help stem cells develop identity Or help relocate cells to final location May “mature” cells into structural type May protect brain cells from injury Synaptic action Copper homeostasis •Anyway, you need it. •Normal “clipping” of APP by a “secretase” enzyme (in red, and also assumed to be a transmembrane protein) is shown. •There are several secretases, also associated proteins, and they seem to mutate easily: there is a genetic link. •It is not exactly clear why things go awry with advanced age. Clipping APP the right & wrong ways NH2 terminus Correct Unlike the α-secretase enzyme, which cuts APP into shorter protein fragments in the cell's cytoplasm, the γ-secretase cleaves its target within the hydrophobic membrane of the cell. The transmembrane proteins called presenilins permit access to the γ site on APP. Mutations in the genes coding for presenilins are frequent causes for autosomal dominant Alzheimer's disease. Incorrect Feature article by Vernon M. Ingram American Scientist on-Line Vol. 91, #4 July-August 2003 http://www.americanscientist.org/template/IssueTOC/issue/394 Exporting the dangerous fragments Figure 6. The Aβ1-42 protein fragment is exported from the cell immediately after being cut from the parent APP molecule. Once it reaches the extracellular space, the peptide refolds to form a sticky shape that clumps together as an insoluble aggregate. Feature article by Vernon M. Ingram American Scientist on-Line Vol. 91, #4 July-August 2003 http://www.americanscientist.org/template/IssueTOC/issue/394 Amyloid hypothesis: fibrils or protofibrils cause cell death, possibly as the body’s own defenses tries to clear such “foreign” matter. Peter Lansbury Group http://focus.hms.harvard.edu/1998/June4_1998/neuro.html Competing hypothesis: channel formation disrupts Ca+2 metabolism Introduction of Varying Salts to Increase β-amyloid Aggregation, Ab 10-35 NaCl NaNO3 10 mm x 10 mm scanning probe microscope images (on mica) of 300 mM Ab10–35 incubated for 8 days at room temperature in 15 mM phosphate buffer containing 50 mM salt. NaF Alzheimer research group, Team goal: Mediate or alter the Aggregation of Ab R = regular H = hydrophobic Exemplify with Double-stick Tape Peptide-based Mediation Requires a Specific Sequence Hydrophobic KLVFF region is responsible for βamyloid aggregation Incorporation of such region for β-sheet breaking or capping H-(Lys)-Val-Leu-Phe-Phe-(Lys)6-NH2 A peptide construct incorporating the KLVFF region developed by Professor Regina Murphy at the University of Wisconsin-Madison Peptide-based Inhibitors of Ab Fibrillogenesis Ab16-20 core Ab16-22 core "Disrupter" H3N H3N H H3N O H N H O N H H O H N H O N H H O O H N 6 H H-Lys-Leu-Val-Phe-Phe-(Lys)6-NH2 "Disrupter" CH3 O H NH2 N H3N O NH3 N H H H O CH3 N H CO2 O N H N O Meredith Ab core analog H2N O H2N H N H NH2 HN H O O N H H N H H O O N H NH3 H HN H2N NH2 H CH3 H N H NH2 O H-Lys-(Me)Leu-Val-(Me)Phe-Phe-(Me)Ala-Glu-NH2 Murphy "Disrupter"? CH3 O H H-DLeu-DPhe-DLeu-DArg-DArg-NH2 Norstedt LSU Peptide-based Mediators K L V F F AMY-1 x = 1, y = 6 AMY-2 x = 6, y = 1 AMY-3 x = 1, y = 1 MCP 1 K L V F F MCP 2 O H2N Dbzg O OH H2N OH Dibg Mediators Developed by Professor Robert Hammer & Professor Mark McLaughlin Synthesis & Vetting of Peptide with aaAA-Blocker KH H2N O H N O V H N iBu iBu H O H N O N Bn Bn H F H O H N Pr Pr O (NH K H NH ) O 6 2 H-Lys-Dibg-Val-Dbzg-Phe-Dpg-(Lys)6-NH2 HPLC of crude peptide MALDI-MS of purified peptide Calc’d for (M + Na) = 1731.3 Determining Mediator Efficacy Using Transmission Electron Microscopy Control 50 mM Ab1-40 4.5 months 10 mM 50 mM Ab1-40 50 mM AMY-1 4.5 months 10 mM 10 mM Control 50 mM Ab1-40 5 mM AMY-1 4.5 months 1:1 Ab:Inhibitor 10:1 Ab:Inhibitor Even a sub-stoichiometric amount of AMY-1 inhibitor is effective TEM image after 4.5 months at Room Temperature 50 mM phosphate buffer/ 150 mM NaCl pH 7.4 But such limited success is very after-the-fact. Can we use diffusion-based and other methods to determine the early stages of aggregation? Can we follow it in real-time in vitro? Two Possible choices: • Dynamic light scattering • Fluorescence photobleaching recovery A series of dynamic light scattering runs can identify a peptide that has an effect on large fibrils. 50000 Addition of 5mL of 500mM Murphy Peptide dissolved in water 40000 b -Amyloid1-40 incubated R h,app / Å 30000 in 100% DMSO followed by dilution in PBS pH 7.4 20000 10000 Sonicate in water bath for 10 mins with probe sonicator 0 0 10000 20000 30000 40000 Time/s 50000 60000 70000 That’s OK for simple screening, but there are problems with DLS 1) the size is only an apparent value, because of the single angle used for measurement; 2) the presence of small protofibrils, and the effect of inhibitors on them, is difficult to ascertain, especially in the presence of larger fibrils that dominate the scattering; 3) reversibility is not easily studied; and, 4) experimentally tedious for early stages of aggregation. Modulation FPR Device Lanni & Ware, Rev. Sci. Instrum. 1982 SCOPE 5-10% bleach depth IF PA c X TA/PVD * PMT * D S * DM OBJ M RR * M ARGON ION LASER AOM * = computer link Labeling β-Amyloid fragment 1-43 H2N-DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIAL-COOH When fluorescein is attached, we call it L-Ab Fluorescein has about 7% the mass of Ab. Sticht, H.; Bayer, P.; Willbold, D.; Dames, S.; Hilbich, C.; Beyreuther, K.; Frank, R.; Rösch, P.Eur. J. Biochem. 1995, 233, 293-298. Diffusion Results – Great Reproducibility But Dye Shrinks it…and may stabilize against aggregation. 6.0 5.5 pH 2.7 pH 6.9 pH 11 Sodium Fluorescein Dye 5.0 4.5 -6 D/10 cm s 2 -1 4.0 3.5 3.0 2.5 2.0 Theoretical/Experimental D0 for monomeric β-amyloid1-40 1.5 1.0 0.5 0.0 0 5 10 15 20 25 30 35 Days 100 μM Mixture β-amyloid1-40 in phosphate buffer – pH 2.7, 6.9 and 11 Theory/Experimental result for monomeric Ab1-40 from: Massi, F.; Peng, J.W.; Lee, J.P.; Straub, J.E. Stimulation Study of the Structure and Dynamics of the Alzheimer’s Amyloid Peptide Congener in Solution. Biophysical Journal 2001, 80, 31-44. Back to DLS: L-Ab does not prevent formation of large fibrils when mixed with unlabeled material and fibrils increase in size with added salt. 100 mM b-Amyloid1-40 in PBS, pH 7.4 7.0 6.5 6.0 5.5 -9 D/10 cm s 2 -1 5.0 4.5 4.0 3.5 3.0 2.5 2.0 1.5 1.0 Mixed labeled & unlabled 0.5 0.0 0 25 50 75 100 125 150 Ionic strength/ mM 175 200 Epifluorescence also shows L-Ab is actually incorporated into macrofibers. Bottom Line: we think L-Ab is OK to study. Two FPR Contrast Decay Modes are Often Observed: Fast = small; Slow = large. Contrast / Arbitrary 1 pH 2.7 pH 6.9 pH 11 0.1 0.01 1E-3 1 10 100 t/s 1000 Doing More Experiments Faster with Less Precious Amyloid: Dialysis FPR Pump Exchange Fluid Cover slip Sample PTFE spacer Dialysis membrane O-ring Evolution of protofibrils from labeled monomer after dialysis against a weak citrate buffer at pH 5.0. After one hour, large aggregates appear and represent ~ 18% of the signal. FPR of 50mM labeled b-amyloid1-40 in 10 mM KOH dialysis againt 10mM Citrate buffer pH 5.0 2.0 -6 Diffusion Coefficient/ 10 cm s 2 -1 2.5 1.5 Amplitude 81.5% ; 82% 1.0 0.5 Amplitude 18.5% ; 18% 0.0 0 20 40 60 80 Time/min 100 120 140 Finding a convenient buffer for controlled self assembly. This run is at pH 4 Acetate Buffer. Adding calcium hastens aggregation. One Pot Dialysis FPR of 50mM 5CFb-Amyloid1-40 in 10mM KOH Slow Fast 2 Exponential Analysis 1E-6 100mM Acetate buffer pH4 0.1N HCl pH1 1E-7 Percent Amplitude Diffusion Coefficient/ cm s 2 -1 50mM PB PH 7.2 0 5 10 15 20 Scan # 1E-8 5mM CaCl2 10mM CaCl2 15mM CaCl2 25mM CaCl2 1E-9 0 2 4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 36 38 40 2 Time/10 mins Fast Slow Amplitudes 100 95 90 85 80 75 70 65 60 55 50 45 40 35 30 25 20 15 10 5 0 25 30 35 Reversing Amyloid Aggregation…by pH FPR Study: Reversibility of b-Amyloid Aggregation 100mM 5-CF-b-Amyloid1-40+ b-Amyloid1-40 pH 11 dialysis against 50mM PB pH 7.4 -6 D/10 cm s 2 -1 1E-6 1E-7 dialysis against 50mM PB pH 2.7 1E-8 0 200 400 600 800 1000 1200 1400 1600 1800 Time/min Diffusion from in situ FPR of 5-carboxyfluorescein-Ab1-40 (25% mixed with unlabeled 75% Ab1-40) starting at pH 11, then alternately dialyzed between 50 mM phosphate (pH 2.7) and 50 mM phosphate (pH 7.4). What has any of this got to do with Nanomaterials??? Ab = $400,000/gram Need cheaper model systems. They also have materials applications. Bolaform amphiphiles have a dumb-bell shape. hydrophilic hydrophilic hydrophobic Arborol example: [9]-10-[9] 9 watery hydroxyl groups OH HO HO H N HO HO HO HO HO HO HO N H O O 10 oily methylene groups O N H O O OH H N OH OH O N H H N OH OH OH OH OH Toolbox Small angle X-ray scattering: analysis of the structure (inverse space) q Basic idea: Detector I Sample Synchrotron I Scattering envelope X-ray q Intensity Melting an 8% Arborol Gel 0.012 pr25 0.01 pr26 pr27 0.008 pr28 0.006 pr29 pr30 0.004 pr32 0.002 pr33 pr34 0 -0.002 pr35 0 0.5 1 1.5 2 q/nm Eleven runs Start time (30oC): 3:35 am End time (90oC): 5:27 am 2.5 pr36 Stacked dumbbell model 1 Based on molecular modeling, SAXS, FF-SEM, DSC, AFM, POM…and common sense. qI(q) 0.1 0.01 1E-3 1E-4 0.0 0.2 0.4 0.6 0.8 1.0 2 -2 1.2 1.4 1.6 q / nm z x I(q) = S(q)P(qr) 1 S(q) 2 M M M j1( j i ) i 1 sin(qrij ) N+N1 3N+(N-1) (back) N+ 2N+ 5 3 3N+5 5 (back) N+ 2N+ 3 1 3N+3 3 (back)N+ 3N+1 1 Origin(0 (back) 4N+2 ,0,0) 2N+ 1 4N+ 1 2N+ 2 2 qrij 3 P(qr ) 3 sin x x cos x x N1 2N+(N1) y 5 3N+2 (back) N+ 2N+ 4 2 3N+4 (back) N+ 2N+ 6 6 3N+6 4 (back)N+ 3 N 6 4N (back) 2 4 N b a 2 Nradius, r Synthesis of Inhibitor [9]-6 O Br bromohexane + O OEt O NaC OEt OEt O DMF/Benzene OEt OEt OEt O O triethyl carbonate OH OH 3 H2 N OH H N OH OH OH O O N H OH DMSO O [9]-6 H N OH OH OH OH OH Self-assembly of [9]-12-[9] Starting point is “extruded” fibers 1300 [9]-10-[9]only [9]-10-[9] plus [9]-6 1200 1100 Rh/(Å) 1000 900 800 Rh from linear fit of gamma vs q2 of DLS data at five angles: 40, 50, 60, 70 and 90. 700 600 500 400 0 1 2 Number of Days 3 4 Scientific Conclusions • Promising inhibitors have been designed and constructed. Probably even more expensive than Ab itself. • DLS can screen promising ones. • Dialysis FPR can observe Ab deconstruction in real time. So far, only by pH, but dialysis experiments with precious inhibitor are coming. • Model systems to practice with can teach us better methods…and have some materials science applications. • Many things not shown: e.g., Ab slows diffusion of the lipids that make cell membranes. Is this important? Broader Conclusions • Membrane proteins (or fragments) are hard to study. • Don’t expect a cure soon and you won’t be disappointed. • Take your statins once the doctor tells you to start, then hope for the best. • Science in the service of practical problems is increasingly multidisciplinary. • Scientists spend a lot more time scratching their heads and wondering what’s going on than it must seem from textbooks. Discussion Points • • • • We have spent $1.4 M for this research (so far). Perhaps 10 papers will appear eventually. About six Ph.D. students will be trained. Could 100,000 teams like ours (that’s 2000 in every state of the United States!) cure Alzheimer’s, Mad Cow, Huntington’s and other related afflictions? Lecture Checkup (do NOT write your name) • What is the single most important thing you learned from this lecture? • What is the ONE thing you wish you understood better about this lecture? • Action item: what will you do to understand better?