Document 14471265
advertisement

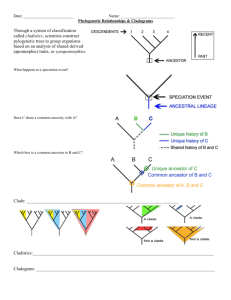
Species evolve with significantly different morphological and behavioural traits due to genetic drift and other selective pressures. Example – Homologous features such as tailbones are evident in the embryos of these organisms. Divergent evolution occurs very rapidly and simultaneously in one species resulting in the production of three or more new species. Ground finches migrated from South America (where they ate medium-sized seeds) to the Galapagos Islands. The island did not exhibit much competition, thus the founding population was very successful. Since there were many food sources available, different seed sizes, insects, and cactus, the ecosystem grew to contain 13 different species which descended from the founding population. Selection patterns operate to produce striking similarities among distantly related species. Some organisms, such as dolphins and shark, are morphologically similar, but their genetic history is different. Both are streamlined, fast swimming, and carnivorous, but a shark is a fish, and a dolphin is a mammal. When traits are similar in appearance, but have different evolutionary origins they are said to be homoplasies (a.k.a. analogous features) Marsupial mammals evolved in isolation from placental mammals, yet in Australia, natural selection favoured species with traits from both groups. One species evolves in response to the evolution of another species – they are dependent on one another for survival (a.k.a. reciprocal adaptation). Yucca moths and yucca plants Yucca flowers are a certain shape so only that tiny moth can pollinate them. The moths lay their eggs in the yucca flowers and the larvae (caterpillars) live in the developing ovary and eat yucca seeds. Recall the classification of living things – Kingdom, Phylum, Class, Order, Family, Genus, Species. This system demonstrates how organisms are anatomically related. A phylogenetic tree demonstrates the evolutionary relationship among organisms. Cladistics is a method for determining the evolutionary histories of organisms – a cladogram is created. The construction is based on the analysis of shared derived (apomorphic) traits, or features that are shared only by members of the group with the common ancestor (these are called synapomorphies). A phylogenetic tree is a cladogram to which additional information, such as evidence of the dates of separation of lineages, has been added. The trunk represents the point in the past when the lineage consisted of only one ancestor. What evidence is used to infer how traits change during evolution? Determine the original state of the trait Determine how the trait has been modified Recall the difference between homologous and analogous features: The supporting structures of bat and bird wings are derived from a common tetrapod (four-limbed) ancestor and are thus homologous. Although they are also used for flight (and thus are analogous), insect wings evolved independently and their supports are not homologous with the wing bones of bats and birds. Consider the following cladogram: A – ancestor to all vertebrates had fins B – ancestor of all non-fish vertebrates had evolved 4 limbs. Remember that ancestral features are not used to determine relationships (eg: presence of a tail) The tree below provides information about the relative sequence in which species diverged or split. The ingroup include species that are chosen for studying. The outgroup are similar, but more distantly related (the first group to diverge from the ingroup). Refer to Table 1 on the handout “Applying Cladistics”. Considering the previous cladogram, which pair of groups is more closely related (i.e.: shares a more recent common ancestor) ? Fern and liverwort or wheat and pine? A: If we trace back the pathways for each of these pairs, they lead to a clade (a point of divergence). A clade represents the point where to groups last shared a common ancestor. Fern and liverwort: diverged at clade B Wheat and pine: diverged at clade D 1. 2. Which group is more closely related to liverwort: moss or maple? A: When we trace back the lineage for moss to see where it diverged from liverwort, and then do the same for maple and liverwort, we see that they diverge from the same point, at clade B. 3. If a different synapomorphy was described and it was known to occur in moss and in pine, in which other groups must it also be found? A: If this trait is seen in pine and moss, there are two possibilities. It might have evolved twice, independently in two groups. Or, it evolved once in history. The most likely of these two possibilities is that it evolved one time, and so must have developed before the divergence of moss. Therefore, all groups after that point must also have the same trait. So we would also see the trait in fern, spruce, maple, wheat and orchid. Amino sequences can be analyzed to construct phylogenetic trees. Eg. Amino acid sequence for cytochrome C, found in mitochondria: Red letters indicate differences from the human sequence while blue letters indicate similarities common to all species studied. Sequences of DNA inserted by viral infection into a species’ genome can be studied to construct phylogeny. SINEs – Short interspersed nuclear elements: repeated DNA sequences of 300 base pairs in length LINEs – Long interspersed nuclear elements: repeated DNA sequences between 5000-7000 bp in length These sequences do not express any RNA or protein Since these insertion events are incredibly rare, if 2 species have the same SINE or LINE located at the same position in their DNA, it is assumed that the insertion occurred only once in a common ancestor.