2/6/2012 Chapter 4 Discovery of the Double Helix Genetics and Cellular Function
advertisement
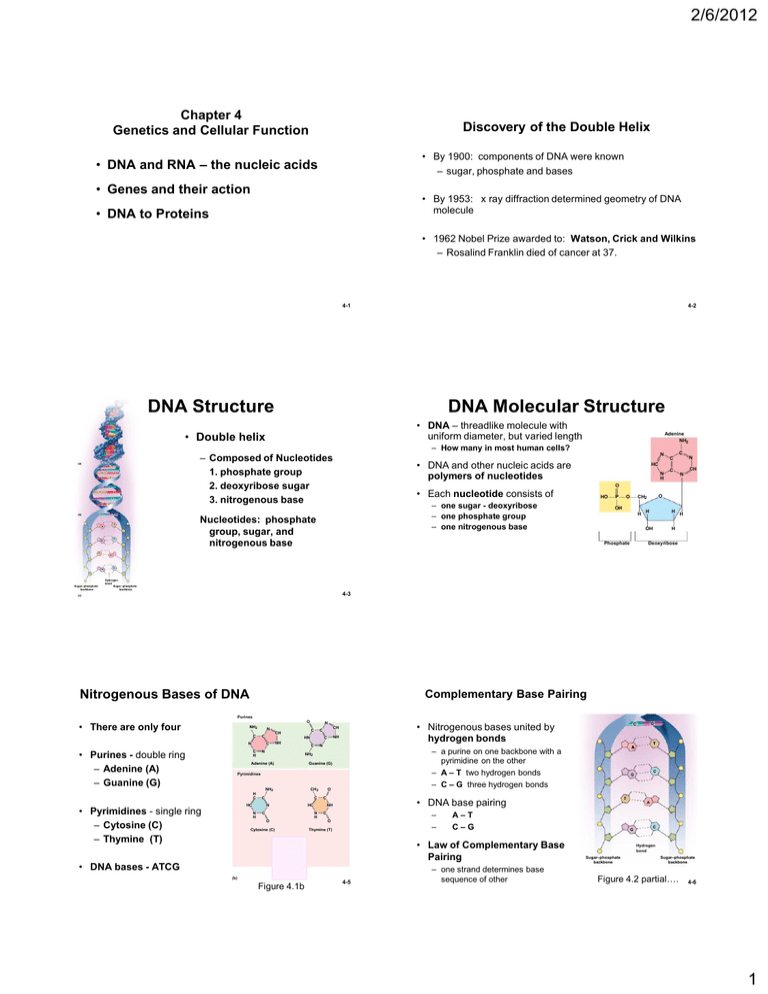
2/6/2012 Chapter 4 Genetics and Cellular Function Discovery of the Double Helix • By 1900: components of DNA were known – sugar, phosphate and bases • DNA and RNA – the nucleic acids • Genes and their action • By 1953: x ray diffraction determined geometry of DNA molecule • DNA to Proteins • 1962 Nobel Prize awarded to: Watson, Crick and Wilkins – Rosalind Franklin died of cancer at 37. 4-1 4-2 DNA Structure DNA Molecular Structure • DNA – threadlike molecule with uniform diameter, but varied length • Double helix (a) A T G C A T A T G C A T C G T A – Composed of Nucleotides 1. phosphate group 2. deoxyribose sugar 3. nitrogenous base T C G N • DNA and other nucleic acids are polymers of nucleotides HC N H C C C N CH N O • Each nucleotide consists of HO – one sugar - deoxyribose – one phosphate group – one nitrogenous base G C (b) Adenine NH2 – How many in most human cells? Nucleotides: phosphate group, sugar, and nitrogenous base P O O CH2 OH H H H H OH Phosphate H Deoxyribose T A G Sugar–phosphate backbone C Hydrogen bond Sugar–phosphate backbone 4-3 (c) Nitrogenous Bases of DNA Complementary Base Pairing Purines O • There are only four NH2 N C C N • Purines - double ring – Adenine (A) – Guanine (G) C H N C CH C HN C NH C N • Nitrogenous bases united by hydrogen bonds CH NH C N Adenine (A) Guanine (G) Pyrimidines H C NH2 • Pyrimidines - single ring – Cytosine (C) – Thymine (T) CH3 C HC C N N H O C HC C O Cytosine (C) – – C O Thymine (T) O HN C • DNA bases - ATCG O (b) C N H CH CH Uracil (U) Figure 4.1b T A–T C–G • Law of Complementary Base Pairing 4-5 – one strand determines base sequence of other C G • DNA base pairing NH N H T – a purine on one backbone with a pyrimidine on the other – A – T two hydrogen bonds – C – G three hydrogen bonds NH2 G C A G C Hydrogen bond Sugar–phosphate backbone Sugar–phosphate backbone Figure 4.2 partial…. 4-6 1 2/6/2012 Chromatin and Chromosomes DNA Function • chromatin – thread of DNA material with proteins – occurs as 46 long filaments called chromosomes – in nondividing cells, chromatin is so slender it cannot be seen – histones – cluster of eight proteins – nucleosome consists of : • Histones with DNA around them • Gene – segment of DNA that codes for a specific protein • Genome - all the genes of one person (or species) – humans have ~ 35,000 • 2% of total DNA • other 98% is non-coding DNA – plays role in chromosome structure – regulation of gene activity – no function at all – “junk” DNA 2 nm 1 DNA double helix 11 nm 2 DNA winds around core particles to form nucleosomes 11 nm in diameter Histone Linker DNA Nucleosome 30 nm 3 300 nm 4 In dividing cells only 700 nm 5 Chromatids Nucleosomes fold accordionlike into zigzag fiber 30 nm fiber is thrown into irregular loops to form a fiber In dividing cells, looped chromatin coils further into a 700 nm fiber to form each chromatid Centromere 700 nm 6 Chromosome at the midpoint (metaphase) of cell division Figure 4.4b 4-7 4-8 What is a Gene? Cells Preparing to Divide • Previous definition - gene - a segment of DNA that carries the code for a particular protein??? • exact copies are made of all DNA (DNA replication) – Body has millions of proteins but only 35,000 genes? – Small % of genes produce only RNA molecules – Some segments of DNA belong to 2 different genes • each chromosome consists of two parallel sister chromatids – joined at centromere – kinetochore – proteins Kinetochore • During cell division - sister chromatids pull apart - each new cell gets a chromatid Centromere • Amino acid sequence of a protein is determined by the nucleotide sequence in the DNA Sister chromatids (a) Figure 4.5a 4-9 4-10 Human Genome Human Genome – Homo sapiens has only about 35,000 genes • Genome – all the DNA in one 23-chromosome set – genes generate millions of different proteins • single gene can code for many different proteins – 3.1 billion nucleotide pairs in human genome • 46 human chromosomes comes in two sets of 23 chromosomes – one set of 23 chromosomes came form each parent – each pair of chromosomes (homologous chromosomes) has same genes but different versions (alleles) exist • Human Genome Project (1990-2003) identified nitrogenous base sequences of 99% of the human genome – genes average about 3,000 bases long • range up to 2.4 million bases – all humans are at least 99.99% genetically identical • 0.01% variations that we can differ from one another in more than 3 million base pairs – some chromosomes are gene-rich and some gene-poor – known locations for >1,400 disease-producing mutations 4-11 4-12 2 2/6/2012 Genetic Code RNA: Structure and Function • Millions of different proteins, all made from 20 amino acids, – encoded by genes made of 4 nucleotides (A,T,C,G) • Genetic code – Arrangement of nucleotides that code for amino acid sequence of proteins i.e., nucleotides arrangement determines amino acid arrangement • Base triplet – a sequence of 3 DNA nucleotides that codes for 1 amino acid – codon - the 3 base sequence in mRNA • RNA – Much smaller than DNA - a single nucleotide chain - Ribose sugar (not deoxyribose) - No thymine nitrogenous base (replaced by Uracil) 3 types of RNA 1. messenger RNA (mRNA) over 10,000 bases 2. ribosomal RNA (rRNA) 3. 3. transfer RNA (tRNA) 70 - 90 bases • Function – interprets code in DNA – uses those instructions for protein synthesis 4-13 Overview of Protein Synthesis • all body cells contain identical genes (except sex cells and some immune cells) 4-14 From DNA to Protein Nuclear envelope • different genes are activated in different cells Transcription • Once activated a gene – Makes messenger RNA (mRNA) – a mirror-image copy of the gene is made DNA Pre-mRNA RNA Processing • migrates from the nucleus to cytoplasm • its code is read by the ribosomes mRNA – ribosomes – ribosomal RNA (rRNA) and enzymes Ribosome – transfer RNA (tRNA) – delivers amino acids to the ribosome Translation – ribosomes assemble amino acids in the order directed by codons of mRNA Polypeptide 4-15 Figure 3.33 Transcription: RNA Polymerase Please note that due to differing operating systems, some animations will not appear until the presentation is viewed in Presentation Mode (Slide Show view). You may see blank slides in the “Normal” or “Slide Sorter” views. All animations will appear after viewing in Presentation Mode and playing each animation. Most animations will require the latest version of the Flash Player, which is available at http://get.adobe.com/flashplayer. • The enzyme that oversees mRNA synthesis • Starts at a promoter site (thanks to a transcription factor) • Breaks H bonds & Unwinds DNA • Adds complementary nucleotides on DNA template strand – following Law of Complimentary Base Pairing – Joins RNA nucleotides together to match DNA coding strand • Encodes a termination signal to stop transcription 3 2/6/2012 Coding strand Termination signal Promoter Template strand Transcription unit In a process mediated by a transcription factor, RNA polymerase binds to promoter and unwinds 16–18 base pairs of the DNA template strand RNA polymerase Unwound DNA RNA polymerase bound to promoter RNA nucleotides mRNA RNA nucleotides RNA polymerase mRNA synthesis begins RNA polymerase moves down DNA; mRNA elongates Please note that due to differing operating systems, some animations will not appear until the presentation is viewed in Presentation Mode (Slide Show view). You may see blank slides in the “Normal” or “Slide Sorter” views. All animations will appear after viewing in Presentation Mode and playing each animation. Most animations will require the latest version of the Flash Player, which is available at http://get.adobe.com/flashplayer. mRNA synthesis is terminated DNA (a) mRNA transcript Coding strand RNA polymerase Unwinding of DNA Rewinding of DNA Template strand RNA nucleotides mRNA RNA-DNA hybrid region (b) 20 Figure 3.34 Fixing pre-mRNA Splicing of mRNA Gene (DNA) • Pre-mRNA is much larger than mRNA – Contains non-coding regions - introns – Coding regions - exons – In nucleus, introns are removed and exons spliced together to produce final mRNA 1 Transcription Intron Pre-mRN A A B C D Exon E F Figure 4.6 2 Splicing mRN A 1 A C mRN A 2 D B D mRN A 3 E A E F 3 Translation Protein 1 • • Protein 2 Protein 3 One gene can code for more than one protein Exons can be spliced together into a variety of different mRNAs. 4-22 3-38 Nucleus From DNA to Protein Nuclear membrane RNA polymerase Nuclear pore Nuclear envelope mRNA Template strand of DNA Transcription Released mRNA DNA Pre-mRNA RNA Processing mRNA Ribosome Translation Polypeptide Figure 3.33 Figure 3.36 4 2/6/2012 Nucleus Nuclear membrane Nucleus Nuclear membrane RNA polymerase Nuclear pore RNA polymerase Nuclear pore mRNA mRNA Template strand of DNA Polysome: mRNA binding to ribosome Template strand of DNA Released mRNA 1 1 After mRNA processing, mRNA leaves nucleus and attaches to ribosome, and translation begins. After mRNA processing, mRNA leaves nucleus and attaches to ribosome, and translation begins. Small ribosomal subunit Codon 15 Codon 16 Codon 17 Amino acids Released mRNA tRNA Aminoacyl-tRNA synthetase Small ribosomal subunit Codon 15 Direction of ribosome advance Portion of mRNA already translated Codon 16 Codon 17 Direction of ribosome advance Portion of mRNA already translated Large ribosomal subunit Large ribosomal subunit Energized by ATP, the correct amino acid is attached to each species of tRNA by aminoacyl-tRNA synthetase enzyme. Figure 3.36 Figure 3.36 Nucleus Nuclear membrane Nucleus Nuclear membrane RNA polymerase Nuclear pore RNA polymerase Nuclear pore mRNA mRNA Template strand of DNA Template strand of DNA Amino acids Released mRNA 1 After mRNA processing, mRNA leaves nucleus and attaches to ribosome, and translation begins. Codon 16 Codon 17 1 tRNA After mRNA processing, mRNA leaves nucleus and attaches to ribosome, and translation begins. Aminoacyl-tRNA synthetase Small ribosomal subunit Codon 15 Amino acids Released mRNA tRNA Aminoacyl-tRNA synthetase Small ribosomal subunit Codon 15 Direction of ribosome advance Portion of mRNA already translated Codon 16 Codon 17 Direction of ribosome advance Portion of mRNA already translated tRNA “head” bearing anticodon tRNA “head” bearing anticodon Large ribosomal subunit 2 Incoming aminoacyltRNA hydrogen bonds via its anticodon to complementary mRNA sequence (codon) at the A site on the ribosome. Large ribosomal subunit Energized by ATP, the correct amino acid is attached to each species of tRNA by aminoacyl-tRNA synthetase enzyme. Energized by ATP, the correct amino acid is attached to each species of tRNA by aminoacyl-tRNA synthetase enzyme. Incoming aminoacyltRNA hydrogen bonds via its anticodon to complementary mRNA sequence (codon) at the A site on the ribosome. 2 As the ribosome moves along the mRNA, a new amino acid is added to the growing protein chain and the tRNA in the A site is translocated to the P site. 3 Figure 3.36 Figure 3.36 Translation of mRNA Nucleus Nuclear membrane RNA polymerase Nuclear pore Cytosol 8 mRNA Nucleus DNA Template strand of DNA Ribosomal subunits rejoin to repeat the process with the same or another mRNA. Amino acids 7 5 Released mRNA mRNA 1 After mRNA processing, mRNA leaves nucleus and attaches to ribosome, and translation begins. Codon 15 Codon 16 Codon 17 4 tRNA 1 Aminoacyl-tRNA synthetase Small ribosomal subunit mRNA leaves the nucleus. 2 Ribosome binds mRNA. Direction of ribosome advance The preceding tRNA hands off the growing protein to the new tRNA, and the ribosome links the new amino acid to the protein. tRNA anticodon binds to complementary mRNA codon. GU Protein A CGU C A U GC 3 Portion of mRNA already translated tRNA “head” bearing anticodon Large ribosomal subunit Once its amino acid is released, tRNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid. 3 As the ribosome moves along the mRNA, a new amino acid is added to the growing protein chain and the tRNA in the A site is translocated to the P site. ADP tRNA 2 4 Incoming aminoacyltRNA hydrogen bonds via its anticodon to complementary mRNA sequence (codon) at the A site on the ribosome. Energized by ATP, the correct amino acid is attached to each species of tRNA by aminoacyl-tRNA synthetase enzyme. A tRNA binds an amino acid; binding consumes 1 ATP. ATP 6 + After translating the entire mRNA, the ribosome dissociates into its two subunits. Pi Free tRNA tRNA is released from the ribosome and is available to pick up a new amino acid and repeat the process. Free amino acids 4-30 Figure 3.36 5 2/6/2012 Please note that due to differing operating systems, some animations will not appear until the presentation is viewed in Presentation Mode (Slide Show view). You may see blank slides in the “Normal” or “Slide Sorter” views. All animations will appear after viewing in Presentation Mode and playing each animation. Most animations will require the latest version of the Flash Player, which is available at http://get.adobe.com/flashplayer. Please note that due to differing operating systems, some animations will not appear until the presentation is viewed in Presentation Mode (Slide Show view). You may see blank slides in the “Normal” or “Slide Sorter” views. All animations will appear after viewing in Presentation Mode and playing each animation. Most animations will require the latest version of the Flash Player, which is available at http://get.adobe.com/flashplayer. 31 Protein Synthesis Information Transfer from DNA to RNA Template strand Figure 3.38 Review of Peptide Formation 3-43 Genetic Code: • RNA codons code for amino acids according to a genetic code • Remember there are 20 amino acids 1 DNA double helix 2 triplets on the template strand of DNA 3 Corresponding codons of mRNA transcribed from the DNA triplets 4 The anticodons of tRNA that bind to the mRNA codons 5 The amino acids carried by those six tRNA molecules 6 The amino acids linked into a peptide chain 4-35 Figure 3.35 6 2/6/2012 Protein Processing and Secretion • Proteins to be secreted are made in ribosomes of rough ER – Contain a leader sequence of 30+ hydrophobic amino acids Inside ER leader sequence is removed; protein is modified • protein synthesis is not finished when the amino acid sequence (primary structure) has been assembled. • to be functional it must coil or folded into precise secondary and tertiary structure • Chaperone proteins (stress proteins or heat-shock proteins) 4-37 3-46 Protein Packaging and Secretion Mechanism of Gene Activation Prolactin Prolactin receptors 1 1 Exocytosis Protein formed by ribosomes on rough ER. ATP 2 Protein packaged into transport vesicle, which buds from ER. Nucleus Casein 7 3 Transport vesicles fuse into clusters that unload protein into Golgi complex. 2 4 Golgi complex modifies protein structure. Secretory vesicles AD P + Pi 6 Golgi complex 5 Golgi vesicle containing finished protein is formed. Regulatory protein (transcription activator) 6 Secretory vesicles release protein by exocytosis. Ribosomes Clathrin-coated transport vesicle Golgi complex Rough Endoplasmic reticulum 5 3 Rough ER 4 Lysosome Casein gene Figure 4.11 mRNA for casein RNA polymerase 4-39 4-40 Synthesizing Compounds Other Than Proteins Please note that due to differing operating systems, some animations will not appear until the presentation is viewed in Presentation Mode (Slide Show view). You may see blank slides in the “Normal” or “Slide Sorter” views. All animations will appear after viewing in Presentation Mode and playing each animation. Most animations will require the latest version of the Flash Player, which is available at http://get.adobe.com/flashplayer. • Cells synthesize glycogen, fat, steroids, phospholipids, pigments, and other compounds – – – – no genes for these synthesis under indirect genetic control produced by enzymatic reactions enzymes are proteins encoded by genes • example – testosterone production – – – – a steroid a cell of the testes takes in cholesterol enzymatically converts it to testosterone only occurs when genes for enzyme are activated 4-42 7







