Genomic Prediction & Selection TRAINING & VALIDATION Agenda
advertisement

6/16/2009 Genomic Prediction & Selection TRAINING & VALIDATION Agenda • A Bioinformatics System to Implement B Bayesian methods for Genomic Prediction i th d f G i P di ti • Results from various methods applied to real and simulated data • Training on EBVs, deregressed data and weighting information in training weighting information in training 1 6/16/2009 Bioinformatics Infrastructure • Research from analysis of high‐density genotypes t to predict merit has several objectives di t it h l bj ti – Determine predictive ability of • same‐density panels in validation/target populations closely related to the training population • same‐density panels in validation/target populations less related or unrelated to the training population related or unrelated to the training population • low‐density panels in populations closely related to the training population – Motivate other genomic selection research Bioinformatics Infrastructure • Identify informative regions for fine‐mapping and gene discovery d di • Provide a platform for collaborating (beef) researchers to undertake genomic training – eg US Meat Animal Research Center • Provide Provide a platform for delivering genomic a platform for delivering genomic predictions to (the beef) industry 2 6/16/2009 Various Methods y = Xb + ∑ Mi a i + e estimate σ ai2 and σ e2 BayesA y = Xb + ∑ Mi a iδ i + e estimate δ i , σ ai2 and σ e2 BayesB estimate δ i , σ a2 and σ e2 BayesC estimate π , δ i , σ a2 and σ e2 BayesCPi Various Methods Markers in Model Marker Effects All (π=0) Fraction (1‐π) Random ‐ Individual Variance (Normal) “Bayes A” (B0) “Bayes B” Random ‐ Constant Var (when in model) Bayes C (C0)=“BLUP” Bayes C Random – Constant Var (when in model) Fraction (1‐π) estimated from data=Bayes CPi Categorical Variants (threshold models) Other Variants (estimate scale, heavy tails) Fixed (LS MSE ) Fixed (LS PRESS) Any π options (Bayes Any π options (Bayes K and Bayes K and Bayes T) StepWise PRESS Bayes A & B are not really truly Bayesian – Bayes C and Bayes Cpi are (Gianola) 3 6/16/2009 Pi influences convergence Priors Influence Shrinkage • Influence varies with method and prior degrees of freedom 4 Bayes A df=4 Bayes A df=3 Bayes A df==4 Bayes A df==3 6/16/2009 9 Bayes B df=4 ∏=0.99 10 5 6/16/2009 Bayes C0 11 Bayes C df=4 ∏=0.99 12 6 6/16/2009 BayesB then BayesA (100 markers) “Heritability” for 100 markers chosen for trait in row, applied to trait in column 0.64 0.50 0.23 0.33 0.29 0.22 0.45 0.30 0.24 0.53 0.61 0.24 0.33 0.29 0.23 0.45 0.30 0.26 0.27 0.29 0.57 0.33 0.29 0.22 0.36 0.30 0.25 0.27 0.27 0.23 0.67 0.29 0.26 0.42 0.30 0.29 0.28 0.24 0.23 0.33 0.57 0.25 0.40 0.35 0.27 0.27 0.29 0.26 0.33 0.29 0.53 0.42 0.30 0.25 0.29 0.29 0.23 0.33 0.29 0.25 0.70 0.26 0.25 0.29 0.27 0.24 0.33 0.29 0.22 0.36 0.63 0.24 0.32 0.27 0.26 0.33 0.29 0.25 0.42 0.30 0.65 13 Bayes B then Bayes A (100 markers) Correlation in training data chosen for trait in row applied to trait in column 0.79 0.68 0.37 0.41 0.42 0.33 0.56 0.46 0.39 0.69 0.76 0.38 0.4 0.44 0.34 0.54 0.42 0.41 0.39 0.41 0.77 0.4 0.39 0.35 0.5 0.4 0.39 0.36 0.36 0.35 0.78 0.41 0.41 0.53 0.45 0.43 0 41 0.41 04 0.4 0 38 0.38 0 36 0.36 0 79 0.79 0 39 0.39 0 51 0.51 0 51 0.51 0 41 0.41 0.39 0.4 0.39 0.45 0.41 0.72 0.55 0.41 0.38 0.41 0.4 0.35 0.45 0.4 0.41 0.87 0.4 0.41 0.43 0.41 0.37 0.4 0.48 0.37 0.5 0.79 0.37 0.44 0.4 0.39 0.44 0.38 0.37 0.5 0.45 0.78 14 7 6/16/2009 1st attempt Cross Validation • Dataset 1 comprising 8 breeds • Select best 100 markers in all data using BayesB g y ✓ B1 ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ ✓ B3 ✓ ✓ B4 ✓ ✓ ✓ B5 ✓ ✓ ✓ ✓ B6 ✓ ✓ ✓ ✓ ✓ B7 ✓ ✓ ✓ ✓ ✓ ✓ B8 ✓ ✓ ✓ ✓ ✓ ✓ ✓ Validation B1 B2 B3 B4 B5 B6 B7 Trraining B2 ✓ B8 Bayes B then Bayes A (100 markers) markers in row chosen from Bayes B on all data, Bayes A trained in cross‐ validation for trait in column, predicting merit in omitted data 0.66 0.53 -0.02 0.09 0.02 -0.06 0.07 0.08 -0.03 0.53 0.65 0.01 0.03 0.1 -0.02 0.06 -0.02 0.06 0.01 0.03 0.68 0.02 -0.03 -0.02 -0.04 -0.01 -0.05 -0.05 -0.06 0.01 0.68 0.02 0.04 0.02 0.08 0.11 0.09 0.07 -0.02 0 0.68 0.04 0 0.2 0.04 -0.02 0.01 0.06 0.14 0.08 0.58 0.11 0.03 -0.03 -0.01 0.01 -0.04 0.14 0 0.1 0.74 -0.07 0.04 0.06 0.05 0.01 0.05 0.22 0.07 0.06 0.69 -0.05 0.08 -0.02 0.02 0.15 -0.08 -0.01 0.01 0.14 0.7 16 8 6/16/2009 StepWise then BayesA 17 StepWise then BayesA Successive datasets have previously best markers removed 18 9 6/16/2009 StepWise and BayesA 19 Proper cross‐validation • Marker subset selection and marker estimation are undertaken on each training ti ti d t k ht i i data subset and used to predict “virgin” data • Correlation dropped to 0.18 (at best) when properly (100 marker subset chosen in training data) cross‐validated g ) 20 10 6/16/2009 Training and Validation Purebred (PB) Purebred (PB) PB Î PB 50K SNP Validation • Almost always SNP that spuriously fit the data well – Having a model that fits the training data well provides relatively little information about how good the prediction will be in new data • Many world‐changing research discoveries are announced in news releases and then never‐to‐be‐ h d f heard‐of‐again i • Training & Validation can be done together to quantify the likely confidence in predictions 11 6/16/2009 Cross Validation Training • Partition the dataset (by sire) into say three groups G1 G2 ✓ G3 ✓ Validation Derive g‐EPD Compute the Compute the correlation between predicted genetic merit from g‐EPD and observed performance G1 Cross Validation Training • Every animal is in exactly one validation set ✓ G1 G2 ✓ G3 ✓ ✓ G1 G2 Validation ✓ ✓ G3 12 6/16/2009 Cross‐Validation • 1800 bulls with EPDs ‐ split into 3 – At random – By sire ID ‐ sire of bulls nested in subset – By sire ID ‐ sires also fitted as fixed effects – By time ‐ oldest, middle‐aged, youngest 25 Results 41028m Random Sire Sire+cg Time Bayes y A (B0) ( ) 0.745 0.726 0.646 0.732 Bayes B (.99) 0.722 0.700 0.618 0.712 Bayes C0 0.746 0.728 0.648 0.730 Bayes C(.50) 0.746 0.728 0.647 0.730 Bayes C(.99) 0.728 0.708 0.625 0.717 C.99/C100m 0.553 0.567 0.389 0.583 StepWise 0.547 0.558 0.393 0.542 PRESS 0.523 0.539 0.365 0.574 100m 26 13 6/16/2009 Simulated SNP Results ‐ 1184 QTL 52566 markers Number of training animals π=0.977 1000 2000 3000 4000 B(true) 0.65 0.76 0.82 0.84 C(true) 0.62 0.74 0.80 0.83 B(inflated) 0.63 0.75 0.80 0.83 C(inflated) 0.60 0.71 0.77 0.80 B(0.50) 0.62 0.74 0.79 0.82 C(0.50) 0.60 0.70 0.75 0.78 B(0) 0.64 0.74 0.79 0.81 C(0) 0.59 0.70 0.75 0.78 True=#QTL/#markers; inflated=0.9 true; heritability=0.5 (Christian Stricker for Swiss Cattle Breeders) 27 Simulated Results 2000 animals Number of QTL 171 493 1184 B(true) 0.88 0.82 0.76 C(true) 0.88 0.81 0.74 B(inflated) 0.84 0.79 0.75 C(inflated) 0.70 0.74 0.71 B(0.50) 0.81 0.78 0.74 C(0 50) C(0.50) 0 65 0.65 0 72 0.72 0 70 0.70 B(0) 0.82 0.77 0.74 C(0) 0.64 0.72 0.70 True=#QTL/#markers; inflated=0.9 true; heritability=0.5 (Christian Stricker for Swiss Cattle Breeders) 28 14 6/16/2009 50k within‐breed predictions 50k within‐breed predictions • These predictions are characterized by correlations between genomic merit and realized correlations between genomic merit and realized performance from 0.5 to 0.7 – They will account for 25 (0.52) to 50% (0.72) genetic variation – Compared to a trait with heritability of 25%, the genomic predictions would be equivalent to observing 6 t 15 ff i i 6 to 15 offspring in a progeny test t t • Correlations of 0.7 are similar to the performance of genomic predictions in dairy cattle 15 6/16/2009 50k within‐breed predictions • These predictions are not as highly accurate as can be achieved in a well designed and b hi d i ll d i d d managed progeny test, say with 100 or more offspring • However, for many traits they are much more reliable for animals of a young age (eg y g g ( gp prior to first selection) than is currently achievable from individual performance Across‐breed prediction • Refers to the process of predicting performance for a breed or cross that was not in the training dataset breed or cross that was not in the training dataset • Critical interest to those selecting breeds that are not well represented in the training populations • May not be as reliable as within‐breed predictions due to complexities associated with non‐additive genetic effects (dominance and epistasis) • Potential can be assessed by simulating the effects of P t ti l b d b i l ti th ff t f major genes using real SNP genotypes on various populations 16 6/16/2009 Introduction • Toosi et al.,(2008) simulated genotypic and phenotypic p yp data – Training in crossbred and MB populations – Successful selection of PB for MB performance • Linkage Disequilibrium (LD) – Simulated LD in pure and MB populations may not accurately reflect real LD in beef cattle populations Objective Training Populations Î Validation Populations Multi‐breed (MB) Purebred (PB) MB Î PB MB Î 50K SNP Purebred (PB) Multi‐breed (MB) PB Î MB 50K SNP 17 6/16/2009 50K SNP Datasets MB Population (N=924) PB Population (N=1086) Angus Angus 239 Brahman 10 Charolais 183 Hereford 78 Limousin 45 1086 Maine-Anjou 137 Shorthorn 97 South Devon 135 Simulation of Additive Genetic Merit and Phenotypic Performance 50K SNP 50K SNP X1 X2 X3 Xk X6 X50K 0, 1 or 2 SNP chosen at random QTL 50, 100, 250, 500 QTL1 QTL2 QTLj X6 Xk xijα j xi1α1 xi 2α 2 Additive Genetic Merit N Phenotypic performance yi = ∑ xijα j + ei j =1 18 6/16/2009 Marker Panels 50K SNP X1 X2 X3 X5 X6 Xk LD=r2 50K w/o QTL X50K Xk HLD1 HLD2 HLDj X5 X1 X2 X3 Xm QTLj X6 HLD50, 100, 250, 500 X50K LD=r2 QTL1 QTL2 QTL 50, 100, 250, 500 Xm Xm X5 N Bayesian Analysis y = 1μ + ∑ x jα jδ j + e j =1 Simulated Phenotypes/real 50k Data • Effect of number of available markers 50 QTL Just QTL QTL + Best markers Q QTL + 50k Train in Multibreed Validate in Purebreed 0.953 0 931 0.931 0.766 Train in Purebreed Validate in Multibreed 0.962 0 938 0.938 0.842 19 6/16/2009 Simulated Phenotypes/real 50k Data • Effect of number of available markers 50 QTL Just QTL QTL + Best markers Q QTL + 50k Just Best markers 50k w/o QTL (real life) Train in Multibreed Train in Purebreed Validate in Purebreed Validate in Multibreed 0.953 0 931 0.931 0.766 0.570 0.388 0.962 0 938 0.938 0.842 0.489 0.422 Kizilkaya et al, ASAS, 2009 Effect of number of available markers • Redundant markers reduce accuracy – Increased type I errors • Accuracy suffers greatly when QTL not on panel – Not enough markers of sufficiently high LD to act as good proxies on a one‐for‐one basis g p • Multibreed population generally inferior to purebred 20 6/16/2009 Purebred or Crossbred Highest LD markers for random QTL with Training in Purebred 1.2 y=1 1.2018x 2018x - 0.371 0 371 R² = 0.48309 Means (r) Purebred = 0.717 Multi-breed = 0.491 1 Multi-Breed (r) 0.8 Few QTL with LD <0.4 in training 0.6 0.4 Many markers erode in validation population 0.2 0 -0.2 0 0.2 0.4 0.6 0.8 1 1.2 Purebred (r) Purebred or Crossbred Highest LD markers for random QTL with Training in Crossbred 1.2 y = 0.8775x + 0.0406 R² = 0.39451 Means (r) Multi-breed = 0.625 Purebred = 0.589 1 Purebred (r) 0.8 0.6 Many QTL with LD <0.4 in training 0.4 Most markers still robust still robust in validation population 0.2 0 -0.2 0 0.2 0.4 0.6 0.8 1 1.2 Multi-Breed (r) 21 6/16/2009 Effect of number of available markers • Easier Easier to find high LD markers in purebreds than to find high LD markers in purebreds than multibreed populations because average LD is higher – Favors the use of purebred populations – Necessitates higher density SNP panels in multibreeds • Markers chosen in purebreds may be less informative in multibreed populations as they will have less LD • Markers that work well in multibreed populations seem p p to work just as well in purebred populations • Nice to have larger multibreed populations & denser panels Correlations between true and predicted genetic merits in validation population Panel: QTL QTL 50 100 250 500 MBÎPB 0.953 0.938 0 840 0.840 0.720 PBÎMB 0.962 0.941 0 853 0.853 0.786 22 6/16/2009 Simulated Phenotypes/real 50k Data • Effect of number of QTL Effect of number of QTL 50k w/o QTL 50 QTL Train in Multibreed Validate in Purebreed 0.388 0.289 0.247 0.200 100 QTL 250 QTL 500 QTL Train in Purebreed Validate in Multibreed 0.422 0.308 0.276 0.299 • Identical trends when panel comprises QTL only • These correlations a/c for < 20% variation at best Correlations between true and predicted genetic merits in validation population Panel: HLD QTL MBÎPB PBÎMB 50 0.570 0.486 100 0.513 0.480 250 0.510 0.429 500 0.372 0.391 23 6/16/2009 Average LD between QTL and HLD marker in PB or MB populations HLD to QTL chosen from PB HLD-QTL LD assessed in PB MB 0.549 0.322 MB 0.412 0.408 Conclusions • MB population – A good choice to carry out genomic selection – Reasonably R bl accurate t estimate ti t off genetic ti merits it of selection candidates in a PB population • Accuracy of genetic merit in genomic selection – Higher with fewer QTL – Erodes when more uninformative SNPs added • The extent of LD hence r2 are highly variable – Lower average r2 in MB than PB populations – No complete LD for all QTL with SNPs – Denser markers are needed 24 6/16/2009 Training and Validation Purebred (PB) Purebred (PB) PB Î PB Reduced Panel Reduced panel within‐breed selection • Two‐stage Bayesian analysis – Run all 50k markers • in each of the three training sets (2&3, 1&3, 1&2) – Select the best 600 markers on model frequency and genomic coverage – Rerun the training and validation analyses using only the markers on the 600 marker panel 25 6/16/2009 50k versus 600 markers 384 SNP Panels • Panels of 600 markers per trait for 8 traits would require a single panel of 4 800 markers would require a single panel of 4,800 markers • Technology is moving such that larger panels are costing the same as smaller panels used to, rather than reducing the cost of smaller panels • Significantly cheaper panels are currently Si ifi tl h l tl limited to 384 (or less) SNP – Allow 100 or so of the best SNP for 3‐4 key traits 26 6/16/2009 Even Smaller Panels Validation in New AI Bulls 27 6/16/2009 Summary – beef cattle in US • 50k 50k within breed (like 5 within breed (like 5‐15 15 progeny) progeny) • 50k across breed (like 1 individual record or 5 progeny) • Reduced panel within breed ( (varies up to 50k accuracy) p y) Validation Statistics • Proportion of additive variation accounted for by the genomic prediction the genomic prediction – Molecular BV used as an observation 1/Multivariate model using the MBV as a trait to estimate (eg ASREML) the genetic correlation 2/ Reduction in estimated sire variance when the MBV is included as a fixed effect in the model 3/ Regression of phenotype on MBV Thallman et al, 2009 BIF 28 6/16/2009 Thallman et al, 2009 BIF BVN BVN res cov estd res cov 0 heritability rg Data Simulated from Additive Model Only 0.1 0.04 0.11 0.08 0.1 0.16 0.21 0.23 0.1 0.36 0.38 0.44 0.1 0.64 0.54 0.64 0.3 0.04 0.06 0.05 0.3 0.16 0.17 0.19 0.3 0.36 0.35 0.40 0.3 0.64 0.64 0.68 0.5 0.04 0.05 0.05 0.5 0.16 0.16 0.18 0.5 0.36 0.35 0.39 0.5 0.64 0.63 0.66 Reduction Regression 0.02 0.17 1.40 0.29 0.04 0.15 0.35 0.66 0.04 0.16 0.36 0.63 0.05 0.21 6.62 -0.23 0.05 0.20 0.42 0.83 0.05 0.18 0.39 0.72 1,000 animals representing 100 sires Training on EBVs 29 6/16/2009 Ideal Model (Equation) g = 1μ + Ma M +ε g is (true) genetic merit (BV) M is columns of covariates (genotypes) a are substitution effects ε is lack-of-fit (hopefully small) Ideal Model g = 1μ + Ma + ε g is genetic merit (BV) var(g) = A? or G? var(Ma) = G genomic relationships var(ε ) = Iσ ε2 ? or cA? for c= σ ε2 σ g2 the fraction of var(g) unaccounted by markers 30 6/16/2009 Towards a Practical Model g + e = 1μ + Ma M + (ε + e ) g is (true) genetic merit (BV) e is usual e (g + e ) is phenotype (no fixed effects) var(ε + e) = cA + Iσ e2 since cov(ε , e') = 0 Or include a random polygenic effects explicitly, with var(u)=A etc Practical Model y = Xb + Ma + (ε + e ) Xb are usual fixed effects var(ε + e) = cA + Iσ e2 Not reasonable to assume ass me var( ar(ε + e) = Iσ e2 unless markers are fitting very well But we do all the time ! And the results are fairly similar 31 6/16/2009 Heterogeneous Residual If y has correlated or heterogeneous residual y = Xb + Ma + (ε + e ) var(ε + e) = cA + R R is the usual value that would be used in MME (eg y is means of varying k numbers of observations) y k = Xb + Ma M + (ε + ek ) Repeatability model σ e2 , var (ek ) = D (diagonal) ⎡⎣ k + (k − 1)rI ⎤⎦ with rI =intraclass correlation of residuals var(ek ) = Heterogeneous Residual (cont) y k = Xb + Ma + (ε + e ) var(ε + e) = cI + D Simplify by ignoring polygenic Then "weights" are the diagonals of R −1 1 c or (arbitrarily) c + var(e ) c + var(e ) so weights i ht approach h 1 as k → ∞ (if rI = 0) r ii = var(e ) = σ e2 ⎡⎣ k + (k − 1)rI ⎤⎦ with rI =intraclass correlation of residuals 32 6/16/2009 Family Data When means are from relatives, relatives rather than the same individuals, genetic relationships can contribute to the intraclass correlation EBVs as data g = Ma + ε g + (ĝ - g) = ĝ = Ma + ε + (ĝ - g) with var( ĝ − g) = PEV > 0 p Similar to previous g + e = Ma + ε + e where var(g+e) > var(g) 33 6/16/2009 EBVs as data g = Ma + ε g + (ĝ - g) = ĝ = Ma + ε + (ĝ - g) with var( ĝ − g) = PEV > 0 Generally var(ĝ − g) = var(g) + var( ĝ) − 2 cov(ĝ, g) p shrinkage g pproperties p But BLUP has special cov(ĝ, g) = var (ĝ) so that var(ĝ − g) = var(g) − var( ĝ) var( ĝ) r2 = ≤ 1 so 0 ≤ var( ĝ) ≤ var(g) var(g) Other Relevant Properties of BLUP cov( ĝ, ĝ − g) = var( ĝ) − cov( ĝ, g) var( ĝ) − var(g) var( ĝ) = 0 = var(g) So prediction errors are uncorrelated with estimated merit ĝ ĝ − g 34 6/16/2009 Other Relevant Properties of BLUP But cov(g, ĝ − g) = cov( ĝ, g) − var(g) = var(g) ( ˆ ) − var(g) ( )<0 Really good animals are underestimated Really bad animals are overestimated g ĝ − g Genomic Prediction usual linear regression of y on (fixed) x cov(y, x) , var(x) (random) regression of EBV on markers ĝ on Ma involves cov(ĝ, Ma) ≈ cov(ĝ, g) for small c But cov(ĝ, g)=var(ĝ) will differ for every animal β y.x = according to its accuracy r 2 35 6/16/2009 Need to “inflate” observations g = Ma + ε g + (kĝ - g) = kĝ = Ma + ε + (kĝ - g) Want to choose k so that cov(g kg cov(g, kĝ - g) = 0 cov(kĝ, g) to be constant Finding k Want cov(g, kĝ - g) = 0 cov(g, kĝ - g) = cov(g, ĝ - g) = kvar(ĝ) - var(g) so we want k = var(g) 1 = var(ĝ) r 2 Want cov(kĝ, g) to be constant cov(kĝ, g)=k var (ĝ)= var(g) var (ĝ)= var(g) var(ĝ) 36 6/16/2009 Implications g ĝ = d, d a deregressed d d "observation" " b " with h h2 = r 2 2 r d It is used in the right-hand side of Cĝ= 2 σe where C is such that the PEV is C−1 = (1 − r 2 )σ g2 Proof d = σ e2 Cĝ= σ e2 (1 − r 2 )σ g2 (1 − h ) ĝ= ĝ 2 ĝ= (1 − r 2 )h 2 r2 More Implications But deregressed observations have heterogeneous variance ε + (kg-g kĝ-g ) with k = r −2 so kr 2 = 1 var (ε +kĝ-g )= var (ε ) + var(kĝ-g) = var (ε ) + k 2 var (ĝ ) + var (g ) − 2k var(ĝ) = var (ε ) + k 2 r 2 var (g ) + var (g ) − 2kr 2 var(g) 1 − r2 r2 Therefore the weights representing diagonals of R -1 are = var (ε ) + (k − 1)var (g ) and k − 1 = w= 1 c + 1 − r2 / r2 ( ) or scaled to w ' = c c + 1 − r2 / r2 ( ) so w' → 1 as r 2 → 1 37 6/16/2009 Removing Parent Average During the deregression process, parent average effects should be removed Why? Animals with own and/or progeny information are shrunk towards the parent average Imagine if many bulls had no own/progeny info y should not contribute anything y g to training g They Imagine if some parents were segregating a major effect We dont want this effect shrunk in all the offspring Deregression is no problem if deregressed information is derived directly from animal models during evaluation Removing Parent Average Deregression and removal of parent average effects can be approximately achieved using only the EBV and r 2 values from trios of the training animal, its sire and dam, by setting up mixed model equations for the parent average and offspring, reconstructing the left-hand side to obtain th published the bli h d reliabilities, li biliti before b f reconstructing t ti the th implied i li d right-hand side to determine the deregressed observation and its appropriate r 2 ignoring the parental contribution 38 6/16/2009 Approximate Method First solve for Z ' Z ⎡ Z ' Z PA + 4 λ ⎢ −2 λ ⎢⎣ −2 λ Z ' Zindividual ⎤ ⎡ ĝPA ⎥⎢ + 2 λ ⎥ ⎢ ĝindividual ⎦⎣ ⎤ ⎡ yPA ⎥=⎢ ⎥⎦ ⎢⎣ yindividual ⎤ ⎥ ⎥⎦ −1 ⎤ ⎡ Z ' Z PA + 4 λ ⎡ C PA ⎤ −2 λ C off ⎢ ⎥ = ⎢ off ⎥ −2 λ Z ' Zindividual + 2 λ ⎥ C individual ⎥⎦ ⎢⎣ ⎢⎣ C ⎦ 2 2 to compute rPA = 0.5 − λC PA and rindividual = 1.0 − λC individual Then use [Z ' Zindividual + λ ][ĝindividual ] = [yindividual ] 2 so rderegressed = 1.0 − λ / (Z ' Zindividual + λ ) Substitution Effects Animal pperspective, p Falconer recognizes g a and d effects as the allelic contributions to phenotypic performance Parent perspective, the allelic contributions to breeding value are α =a + d(q - p) Mixed animal and offspring data will be estimating a mixture of a and α effects 39
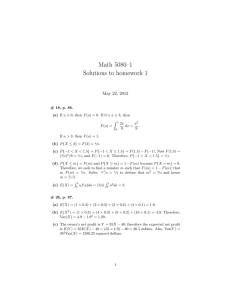
![Problem sheet 1 (a) E[aX + b] = aE[X] + b](http://s2.studylib.net/store/data/012919538_1-498bfd427f243c5abfa36cc64f89d9e7-300x300.png)