6.4 – DNA Replication and Repair Recall:
advertisement

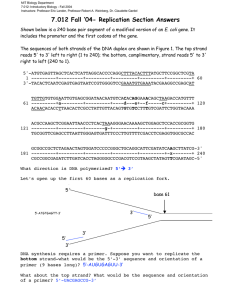
1 6.4 – DNA Replication and Repair Recall: Nucleotide base pairing – purines and pyrimidines. 5’ --> 3’ H-Bonds join complementary bases. Sugar-phosphate backbone. ____ASE = enzyme Why does DNA need to replicate? At what specific point in the cell cycle does DNA replication occur? Conservative vs. Semiconservative Replication Conservative – Copy the entire DNA molecule as it is. The parent double helix strands remain together. Semiconservative – The parent double helix is separated into its 2 strands and a new complementary strand is created for each parent strand. 2 Matthew Meselson and Franklin Stahl – 1958 experiments with DNA using nitrogen isotopes. DNA does in fact replicate semiconservatively! Figure 1 (p.282) 3 The DNA Replication Process DNA replication occurs in 3 steps: Step 1: Strand Separation In order to replicate semiconservatively, the DNA double helix must unwind and break apart its two strands. This will occur at a specific starting point on the DNA called the replication origin. DNA helicase will bind to a replication origin and begin to break apart the hydrogen bonds between base pairs; effectively unwinding the two strands. The unwinding of the two strands creates a replication fork at the point of separation. Unwinding of the strands creates tension that must be overcome if the DNA is to continue separation. Topoisomerase will cleave one or two DNA strands, relieving tension and allowing them to untwist. Topoisomerase will also rejoin the cleaved strands after. Once the strands are separated, they have a tendency to want to rejoin or anneal. Why? Single-strand binding proteins (SSB) will attach to the separated DNA strands and stabilize them to prevent annealing and maintain separation. 4 Figure 3 (p.284) DNA helicase is able to separate the DNA strands in both directions of a replication origin. This means that the new complementary nucleotides are added to both ends as the strand is replicated in opposite directions. The result is multiple replication bubbles which will continue to expand until they meet each other and merge into a full, new DNA strand. 5 Figure 4 (p.284) Eukaryotes can replicate DNA at a rate of 50 base pairs per second at a replication fork. Because of the long length of each DNA strand, this would mean reproducing the entire human genome would take roughly 1 month to complete. However, we know that multiple replication origins and replication bubbles are used at the same time, making the process much more efficient at about 1 hour. Step 2: Building Complementary Strands The DNA strand is built by enzymes called DNA polymerases. Their function is limited to only being able to add new nucleotides to the 3’ end of the strand. Therefore, the DNA is always built from the 5’ end towards the 3’ end. How the nucleotides are actually assembled into the growing strand: The piece that the polymerase attaches to the new strand is actually a nucleoside triphosphate (nitrogenous base + deoxyribose sugar + 3 phosphates). Along with 6 being a building block, a nucleoside triphosphate provides its own energy for the process in the form of the two extra phosphates. Hydrolysis of 2 phosphates and the formation of a phosphodiester bond between the remaining phosphate and the sugar of the previous nucleotide on the strand (sugar-phosphate backbone is formed). Figure 5 (p.285) When a replication fork is opened, nucleotides cannot just simply be added on to build the strand. A small piece of complementary RNA, called an RNA primer, must first be added on to the beginning of the strand. The RNA primer is built by RNA primase. 7 Once the RNA primer is in place, DNA polymerase III will add nucleotides to the RNA primer. Remember that the process is semiconservative. This means that one of the split strands will be oriented 5’ to 3’ and the other 3’ to 5’. However, also recall that DNA polymerases can only add 5’ to 3’. Therefore, the complementary new strands are built in opposite directions. Consequence: As the double helix further unwinds: The new strand that is built towards the fork can be done continuously. We call this the leading strand. The other strand that is built away from the fork opening can only be assembled chunk by chunk (not continuously) when enough fork has opened. We call this the lagging strand. Figure 6 (p.287) 8 Because of the discontinuous assembly on the lagging strand, an extra RNA primer must be placed each time a chunk is to be replicated. The result is multiple short pieces of RNA primer followed by some DNA. We call these segments Okazaki fragments. About 100-200 base pairs long in eukaryotes and 1000-2000 in prokaryotes. Eventually, the next chunk being replicated hits the Okazaki fragment of the piece that was just made before it. We cannot have RNA present in our replicated DNA strand, so DNA polymerase I will remove the RNA nucleotides and replace it with the proper DNA equivalent. Finally, DNA ligase will catalyze the formation of the phosphodiester bonds between all of the nucleotides on the newly created strands. 9 Step 3: Fixing Errors The assembly of the new DNA strands is sadly not a perfectly executed process. What type of errors can occur? DNA polymerase II is used to repair damage to DNA. Figure 7 (p.288) Read 6.4 (pages 282-290). Answer questions #1-4, 6-10 (page 290).